Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Hildebrandt, J., Gorls, H., Hafner, N., Ferraro, G., Durst, M., Runnebaum, I.B., Weigand, W., Merlino, A.(2016) Dalton Trans 45: 12283-12287
- PubMed: 27427335
- DOI: https://doi.org/10.1039/c6dt02380k
- Primary Citation of Related Structures:
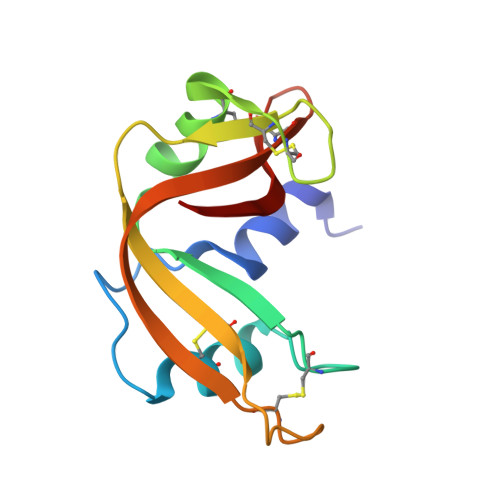
5JLG - PubMed Abstract:
A new pseudo-octahedral π-arene ruthenium(ii) piano-stool compound, containing an O,S-bidentate ligand (compound 1) and showing significant cytotoxic activity in vitro, was synthesized and characterized. In solution stability and interaction with the model protein bovine pancreatic ribonuclease (RNase A) were investigated by using UV-Vis absorption spectroscopy. Its crystal structure and that of the adduct formed upon reaction with RNase A were obtained by X-ray crystallography. The comparison between the structure of purified compound 1 and that of the fragment bound to RNase A reveals an unusual mode of protein binding that includes ligand exchange and alteration of coordination sphere geometry.
- Department of Inorganic and Analytical Chemistry, University of Jena, Germany. wolfgang.weigand@uni-jena.de.
Organizational Affiliation: