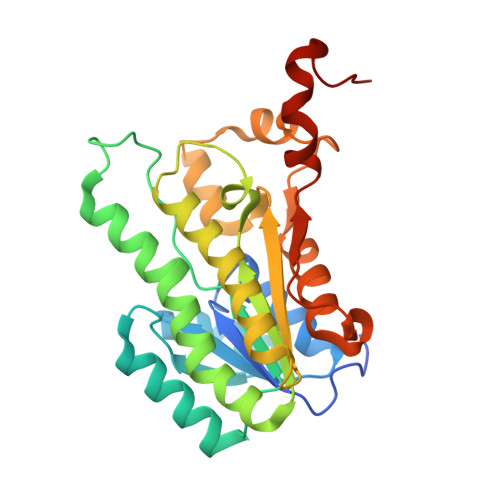
Crystal Structure of putative short-chain dehdrogenase/reductase from Burkholderia xenovorans LB400 bound to NAD
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Delker, S.L., Abendroth, J., Lorimer, D., Edwards, T.E.To be published.