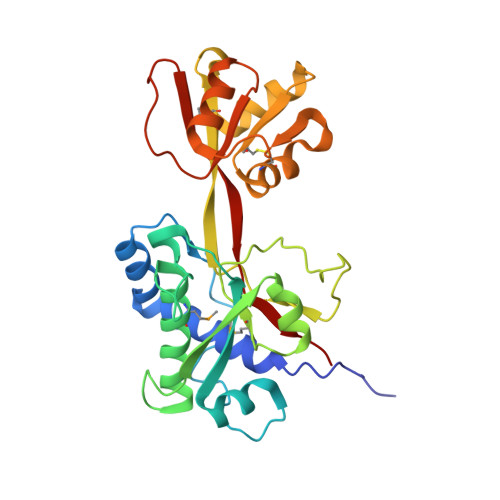
The structure of Plasmodium falciparum 3D7_0606800 reveals a bi-lobed architecture that supports re-annotation as a Venus Flytrap protein.
Parker, M.L., Ramaswamy, R., van Gordon, K., Powell, C.J., Bosch, J., Boulanger, M.J.(2017) Protein Sci 26: 1878-1885
- PubMed: 28681555
- DOI: https://doi.org/10.1002/pro.3218
- Primary Citation of Related Structures:
5JKQ - PubMed Abstract:
Plasmodium falciparum, the causative agent of malaria, employs a diverse array of surface displayed proteins to promote dissemination and establish infection in the human host. Of these, Pf3D7_0606800 is highly immunogenic and has been designated a potential top 10 candidate for inclusion in a multicomponent malarial vaccine. The role of Pf3D7_0606800 in parasite biology, however, is unknown and its characterization has been complicated by a lack of sequence identity with proteins of known structure or function. Towards elucidating Pf3D7_0606800 function, we determined its structure to a resolution of 2.35 Å using selenium single wavelength anomalous dispersion. A bi-lobed architecture displays the core structural hallmarks of Venus Flytrap (VFT) proteins prompting us to re-annotate Pf3D7_0606800 as PfVFT1. Structural analysis further revealed an extended inter-lobe groove that, when interrogated by molecular docking, appears well suited to bind peptide-based ligands. Collectively, our structural characterization of the highly antigenic P. falciparum surface protein PfVFT1 provides intriguing functional insight and establishes a structural template that could prove valuable for malaria vaccine engineering studies.
- Biochemistry & Microbiology, University of Victoria, Victoria, British Columbia, V8W 3P6, Canada.
Organizational Affiliation: