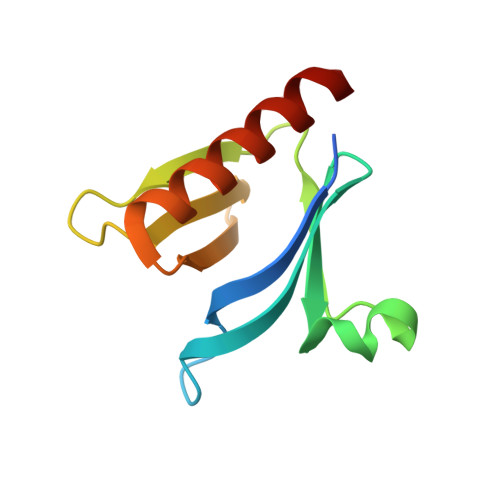
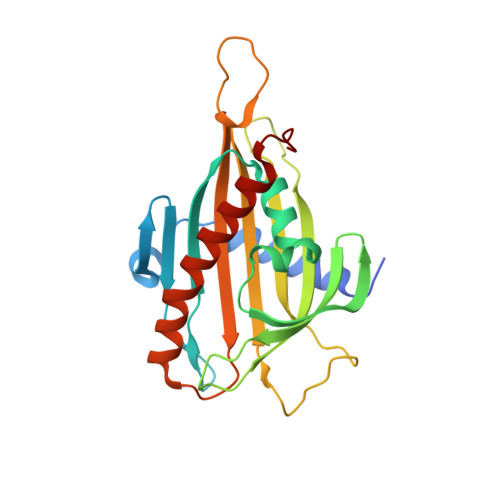
Interaction between the PH and START domains of ceramide transfer protein competes with phosphatidylinositol 4-phosphate binding by the PH domain.
Prashek, J., Bouyain, S., Fu, M., Li, Y., Berkes, D., Yao, X.(2017) J Biological Chem 292: 14217-14228
- PubMed: 28652409
- DOI: https://doi.org/10.1074/jbc.M117.780007
- Primary Citation of Related Structures:
5JJD - PubMed Abstract:
De novo synthesis of the sphingolipid sphingomyelin requires non-vesicular transport of ceramide from the endoplasmic reticulum to the Golgi by the multidomain protein ceramide transfer protein (CERT). CERT's N-terminal pleckstrin homology (PH) domain targets it to the Golgi by binding to phosphatidylinositol 4-phosphate (PtdIns(4)P) in the Golgi membrane, whereas its C-terminal StAR-related lipid transfer domain (START) carries out ceramide transfer. Hyperphosphorylation of a serine-rich motif immediately after the PH domain decreases both PtdIns(4)P binding and ceramide transfer by CERT. This down-regulation requires both the PH and START domains, suggesting a possible inhibitory interaction between the two domains. In this study we show that isolated PH and START domains interact with each other. The crystal structure of a PH-START complex revealed that the START domain binds to the PH domain at the same site for PtdIns(4)P-binding, suggesting that the START domain competes with PtdIns(4)P for association with the PH domain. We further report that mutations disrupting the PH-START interaction increase both PtdIns(4)P-binding affinity and ceramide transfer activity of a CERT-serine-rich phosphorylation mimic. We also found that these mutations increase the Golgi localization of CERT inside the cell, consistent with enhanced PtdIns(4)P binding of the mutant. Collectively, our structural, biochemical, and cellular investigations provide important structural insight into the regulation of CERT function and localization.
- From the Division of Molecular Biology and Biochemistry, School of Biological Sciences, University of Missouri-Kansas City, Kansas City, Missouri 64110.
Organizational Affiliation: