The Structure of Treponema pallidum Tp0624 Reveals a Modular Assembly of Divergently Functionalized and Previously Uncharacterized Domains.
Parker, M.L., Houston, S., Wetherell, C., Cameron, C.E., Boulanger, M.J.(2016) PLoS One 11: e0166274-e0166274
- PubMed: 27832149
- DOI: https://doi.org/10.1371/journal.pone.0166274
- Primary Citation of Related Structures:
5JIR - PubMed Abstract:
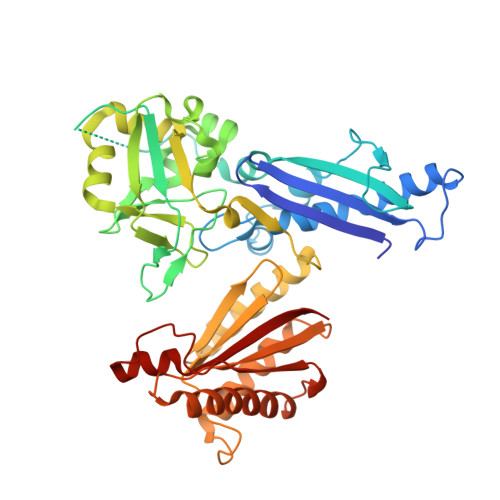
Treponema pallidum subspecies pallidum is the causative agent of syphilis, a chronic, multistage, systemic infection that remains a major global health concern. The molecular mechanisms underlying T. pallidum pathogenesis are incompletely understood, partially due to the phylogenetic divergence of T. pallidum. One aspect of T. pallidum that differentiates it from conventional Gram-negative bacteria, and is believed to play an important role in pathogenesis, is its unusual cell envelope ultrastructure; in particular, the T. pallidum peptidoglycan layer is chemically distinct, thinner and more distal to the outer membrane. Established functional roles for peptidoglycan include contributing to the structural integrity of the cell envelope and stabilization of the flagellar motor complex, which are typically mediated by the OmpA domain-containing family of proteins. To gain insight into the molecular mechanisms that govern peptidoglycan binding and cell envelope biogenesis in T. pallidum we report here the structural characterization of the putative OmpA-like domain-containing protein, Tp0624. Analysis of the 1.70 Å resolution Tp0624 crystal structure reveals a multi-modular architecture comprised of three distinct domains including a C-terminal divergent OmpA-like domain, which we show is unable to bind the conventional peptidoglycan component diaminopimelic acid, and a previously uncharacterized tandem domain unit. Intriguingly, bioinformatic analysis indicates that the three domains together are found in all orthologs from pathogenic treponemes, but are not observed together in genera outside Treponema. These findings provide the first structural insight into a multi-modular treponemal protein containing an OmpA-like domain and its potential role in peptidoglycan coordination and stabilization of the T. pallidum cell envelope.
- Department of Biochemistry & Microbiology, University of Victoria, Victoria, British Columbia, Canada.
Organizational Affiliation: