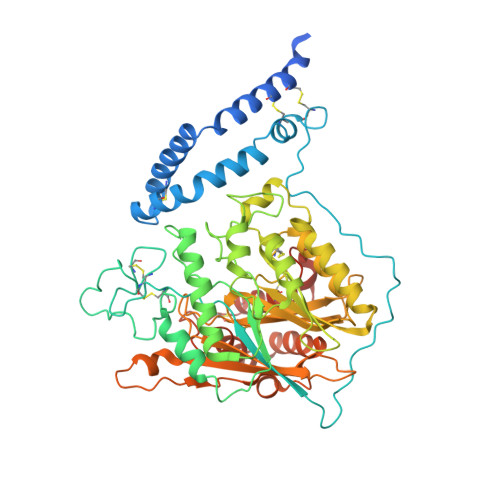
Structure of Human Acid Sphingomyelinase Reveals the Role of the Saposin Domain in Activating Substrate Hydrolysis.
Xiong, Z.J., Huang, J., Poda, G., Pomes, R., Prive, G.G.(2016) J Mol Biology 428: 3026-3042
- PubMed: 27349982
- DOI: https://doi.org/10.1016/j.jmb.2016.06.012
- Primary Citation of Related Structures:
5JG8 - PubMed Abstract:
Acid sphingomyelinase (ASM) is a lysosomal phosphodiesterase that catalyzes the hydrolysis of sphingomyelin to produce ceramide and phosphocholine. While other lysosomal sphingolipid hydrolases require a saposin activator protein for full activity, the ASM polypeptide incorporates a built-in N-terminal saposin domain and does not require an external activator protein. Here, we report the crystal structure of human ASM and describe the organization of the three main regions of the enzyme: the N-terminal saposin domain, the proline-rich connector, and the catalytic domain. The saposin domain is tightly associated along an edge of the large, bowl-shaped catalytic domain and adopts an open form that exposes a hydrophobic concave surface approximately 30Å from the catalytic center. The calculated electrostatic potential of the enzyme is electropositive at the acidic pH of the lysosome, consistent with the strict requirement for the presence of acidic lipids in target membranes. Docking studies indicate that sphingomyelin binds with the ceramide-phosphate group positioned at the binuclear zinc center and molecular dynamic simulations indicate that the intrinsic flexibility of the saposin domain is important for monomer-dimer exchange and for membrane interactions. Overall, ASM uses a combination of electrostatic and hydrophobic interactions to cause local disruptions of target bilayers in order to bring the lipid headgroup to the catalytic center in a membrane-bound reaction.
- Department of Biochemistry, University of Toronto, Toronto, Ontario, Canada.
Organizational Affiliation: