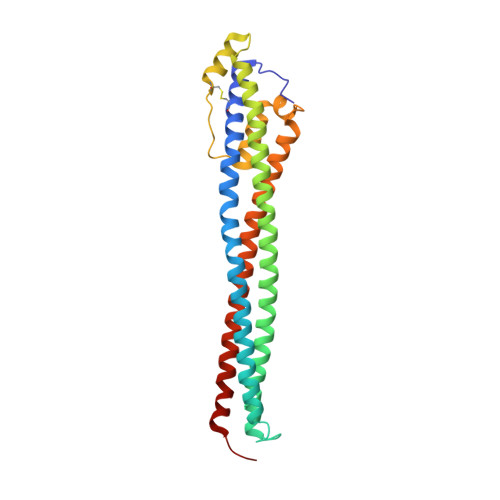
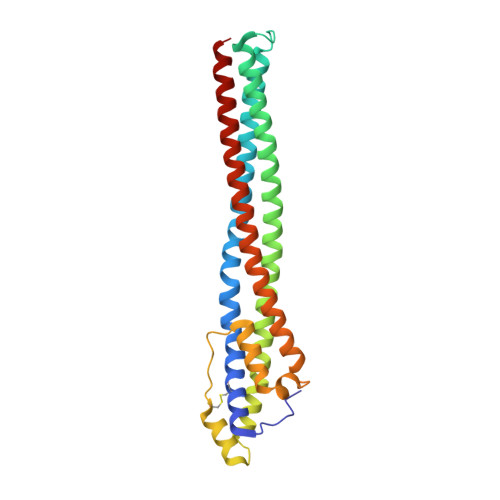
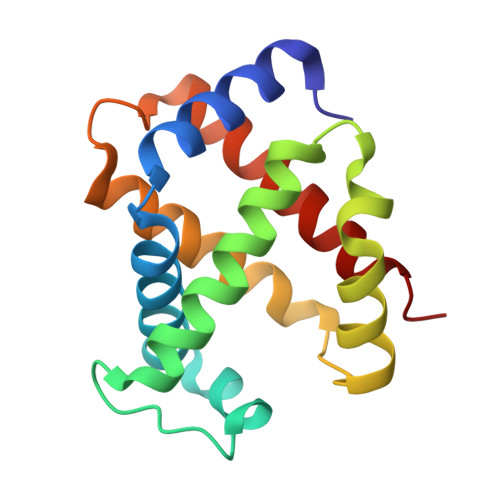
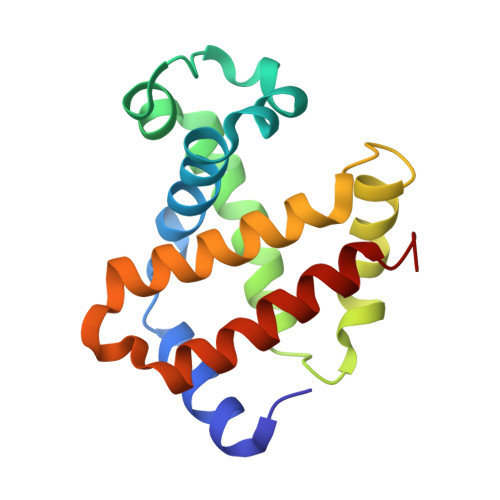
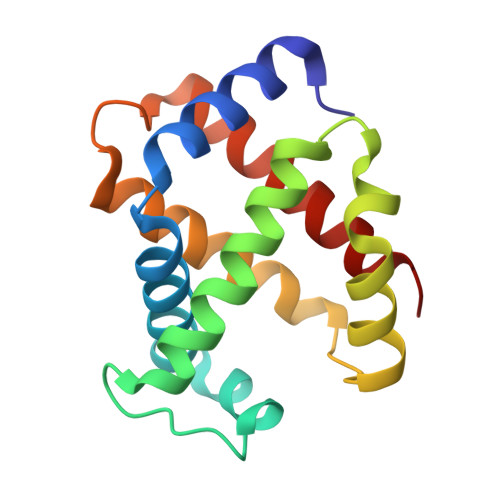
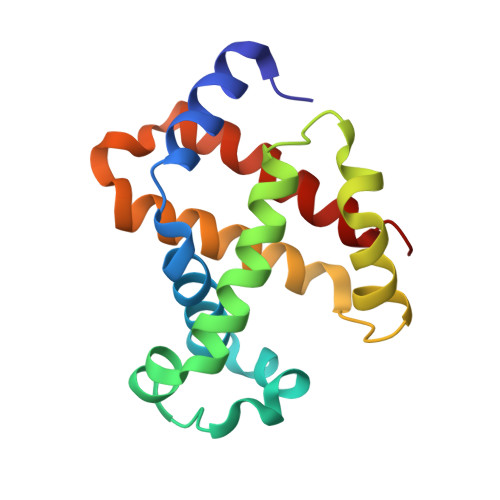
Evolutionary diversification of the trypanosome haptoglobin-haemoglobin receptor from an ancestral haemoglobin receptor.
Lane-Serff, H., MacGregor, P., Peacock, L., Macleod, O.J., Kay, C., Gibson, W., Higgins, M.K., Carrington, M.(2016) Elife 5
- PubMed: 27083048
- DOI: https://doi.org/10.7554/eLife.13044
- Primary Citation of Related Structures:
5JDO - PubMed Abstract:
The haptoglobin-haemoglobin receptor of the African trypanosome species, Trypanosoma brucei, is expressed when the parasite is in the bloodstream of the mammalian host, allowing it to acquire haem through the uptake of haptoglobin-haemoglobin complexes. Here we show that in Trypanosoma congolense this receptor is instead expressed in the epimastigote developmental stage that occurs in the tsetse fly, where it acts as a haemoglobin receptor. We also present the structure of the T. congolense receptor in complex with haemoglobin. This allows us to propose an evolutionary history for this receptor, charting the structural and cellular changes that took place as it adapted from a role in the insect to a new role in the mammalian host.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: