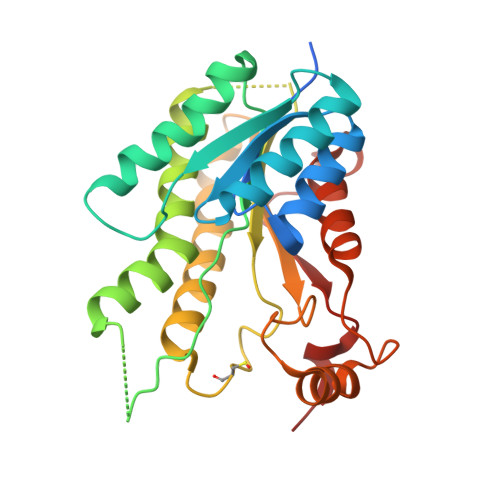
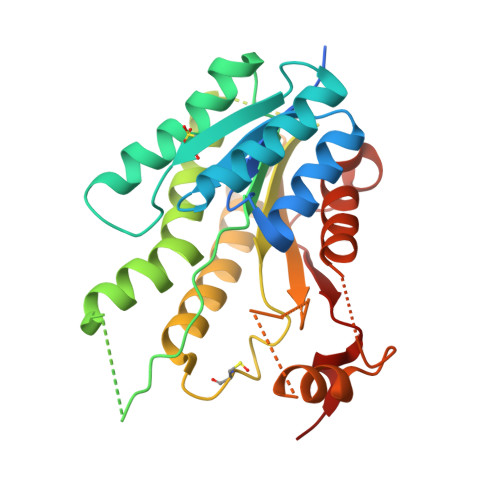
Profiling of Flavonol Derivatives for the Development of Antitrypanosomatidic Drugs.
Borsari, C., Luciani, R., Pozzi, C., Poehner, I., Henrich, S., Trande, M., Cordeiro-da-Silva, A., Santarem, N., Baptista, C., Tait, A., Di Pisa, F., Dello Iacono, L., Landi, G., Gul, S., Wolf, M., Kuzikov, M., Ellinger, B., Reinshagen, J., Witt, G., Gribbon, P., Kohler, M., Keminer, O., Behrens, B., Costantino, L., Tejera Nevado, P., Bifeld, E., Eick, J., Clos, J., Torrado, J., Jimenez-Anton, M.D., Corral, M.J., Alunda, J.M., Pellati, F., Wade, R.C., Ferrari, S., Mangani, S., Costi, M.P.(2016) J Med Chem 59: 7598-7616
- PubMed: 27411733
- DOI: https://doi.org/10.1021/acs.jmedchem.6b00698
- Primary Citation of Related Structures:
5JCJ, 5JCX, 5JDC, 5JDI - PubMed Abstract:
Flavonoids represent a potential source of new antitrypanosomatidic leads. Starting from a library of natural products, we combined target-based screening on pteridine reductase 1 with phenotypic screening on Trypanosoma brucei for hit identification. Flavonols were identified as hits, and a library of 16 derivatives was synthesized. Twelve compounds showed EC50 values against T. brucei below 10 μM. Four X-ray crystal structures and docking studies explained the observed structure-activity relationships. Compound 2 (3,6-dihydroxy-2-(3-hydroxyphenyl)-4H-chromen-4-one) was selected for pharmacokinetic studies. Encapsulation of compound 2 in PLGA nanoparticles or cyclodextrins resulted in lower in vitro toxicity when compared to the free compound. Combination studies with methotrexate revealed that compound 13 (3-hydroxy-6-methoxy-2-(4-methoxyphenyl)-4H-chromen-4-one) has the highest synergistic effect at concentration of 1.3 μM, 11.7-fold dose reduction index and no toxicity toward host cells. Our results provide the basis for further chemical modifications aimed at identifying novel antitrypanosomatidic agents showing higher potency toward PTR1 and increased metabolic stability.
- Department of Life Sciences, University of Modena and Reggio Emilia , Via G. Campi 103, 41125 Modena, Italy.
Organizational Affiliation: