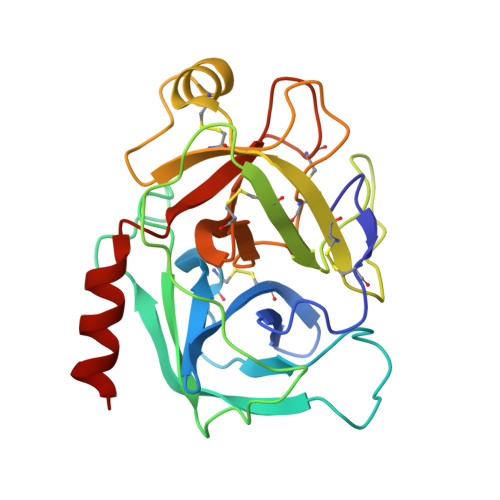
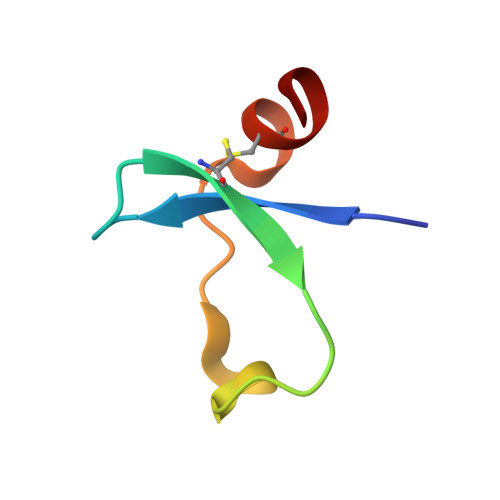
An Acrobatic Substrate Metamorphosis Reveals a Requirement for Substrate Conformational Dynamics in Trypsin Proteolysis.
Kayode, O., Wang, R., Pendlebury, D.F., Cohen, I., Henin, R.D., Hockla, A., Soares, A.S., Papo, N., Caulfield, T.R., Radisky, E.S.(2016) J Biological Chem 291: 26304-26319
- PubMed: 27810896
- DOI: https://doi.org/10.1074/jbc.M116.758417
- Primary Citation of Related Structures:
5JBT - PubMed Abstract:
The molecular basis of enzyme catalytic power and specificity derives from dynamic interactions between enzyme and substrate during catalysis. Although considerable effort has been devoted to understanding how conformational dynamics within enzymes affect catalysis, the role of conformational dynamics within protein substrates has not been addressed. Here, we examine the importance of substrate dynamics in the cleavage of Kunitz-bovine pancreatic trypsin inhibitor protease inhibitors by mesotrypsin, finding that the varied conformational dynamics of structurally similar substrates can profoundly impact the rate of catalysis. A 1.4-Å crystal structure of a mesotrypsin-product complex formed with a rapidly cleaved substrate reveals a dramatic conformational change in the substrate upon proteolysis. By using long all-atom molecular dynamics simulations of acyl-enzyme intermediates with proteolysis rates spanning 3 orders of magnitude, we identify global and local dynamic features of substrates on the nanosecond-microsecond time scale that correlate with enzymatic rates and explain differential susceptibility to proteolysis. By integrating multiple enhanced sampling methods for molecular dynamics, we model a viable conformational pathway between substrate-like and product-like states, linking substrate dynamics on the nanosecond-microsecond time scale with large collective substrate motions on the much slower time scale of catalysis. Our findings implicate substrate flexibility as a critical determinant of catalysis.
- From the Departments of Cancer Biology and.
Organizational Affiliation: