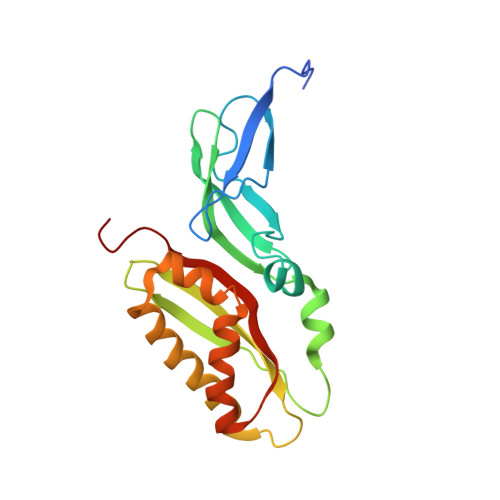
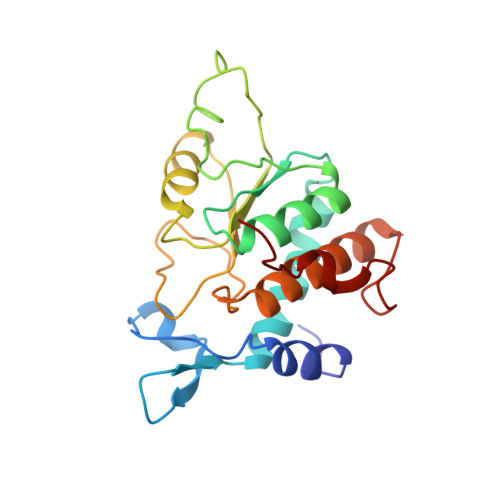
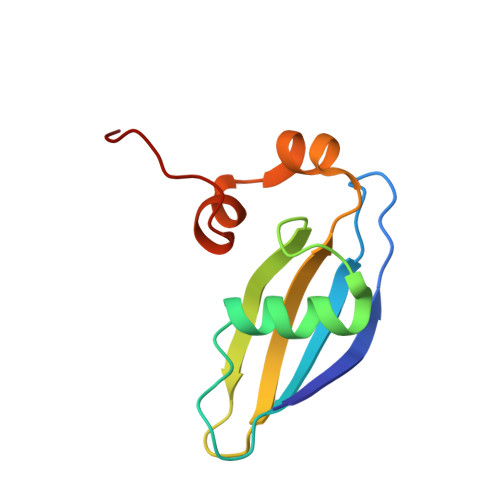
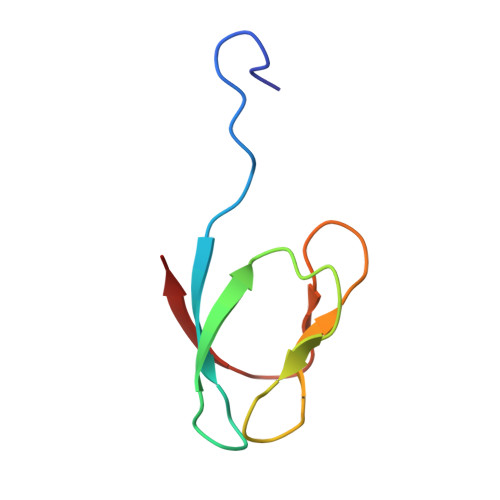
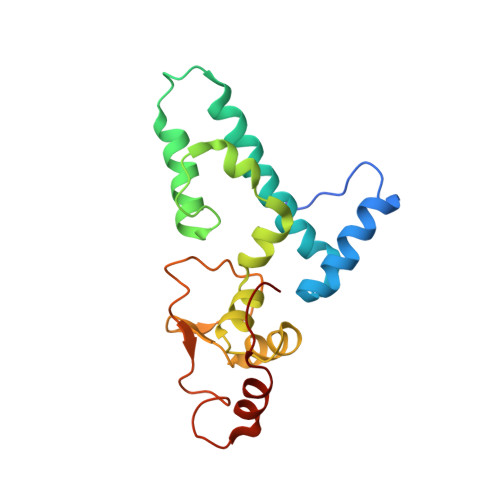
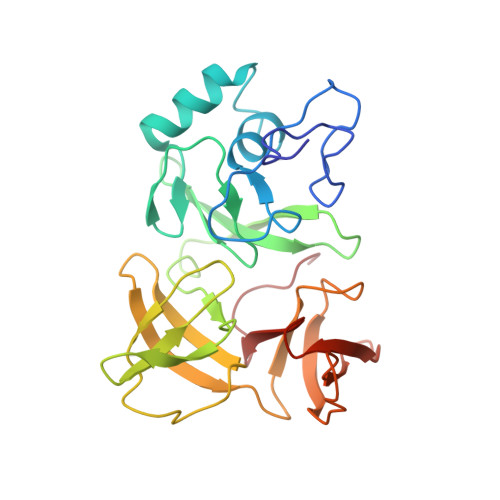
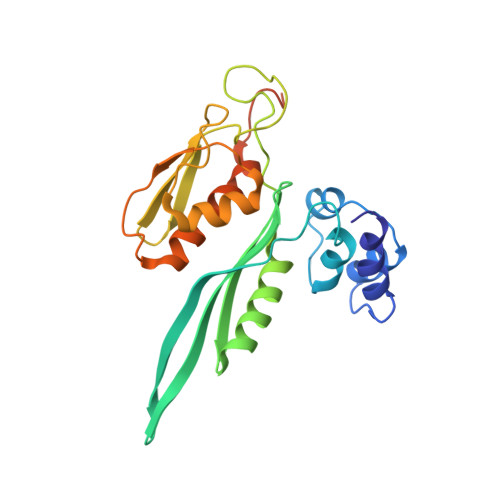
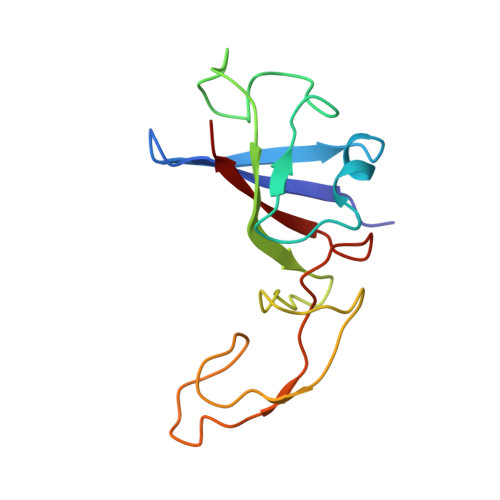
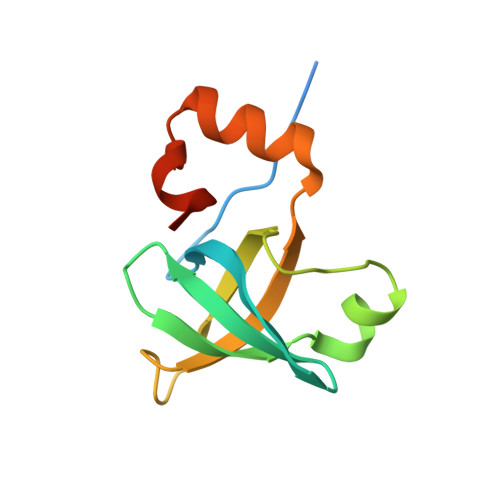
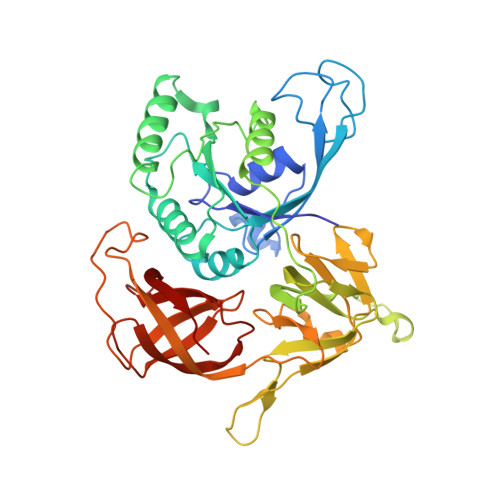
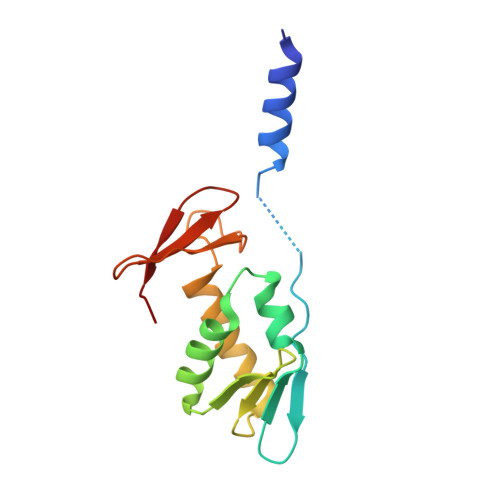
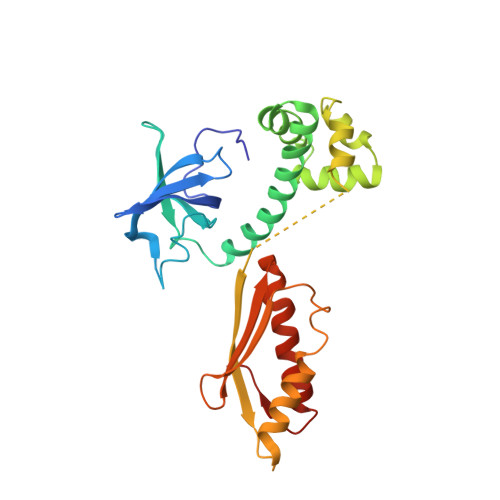
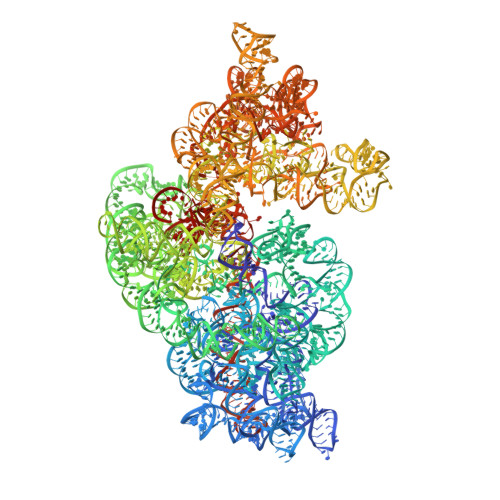
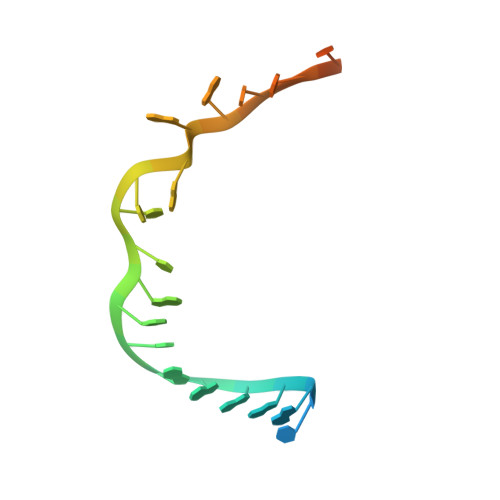
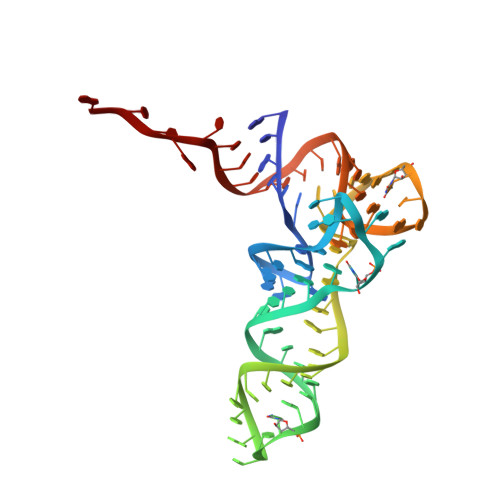
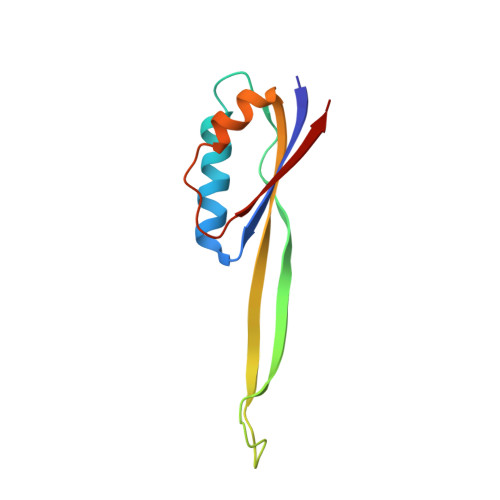Cryo-EM study of start codon selection during archaeal translation initiation.
Coureux, P.D., Lazennec-Schurdevin, C., Monestier, A., Larquet, E., Cladiere, L., Klaholz, B.P., Schmitt, E., Mechulam, Y.(2016) Nat Commun 7: 13366-13366
- PubMed: 27819266
- DOI: https://doi.org/10.1038/ncomms13366
- Primary Citation of Related Structures:
5JB3, 5JBH - PubMed Abstract:
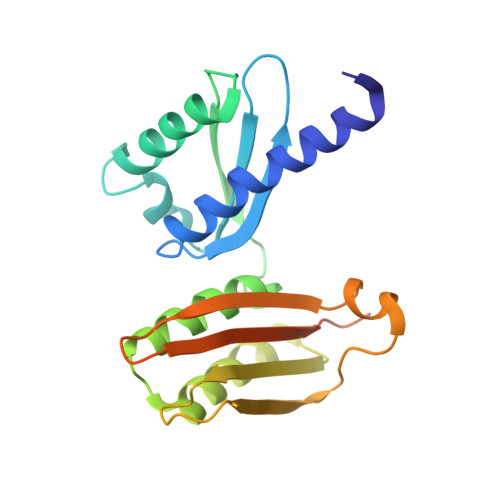
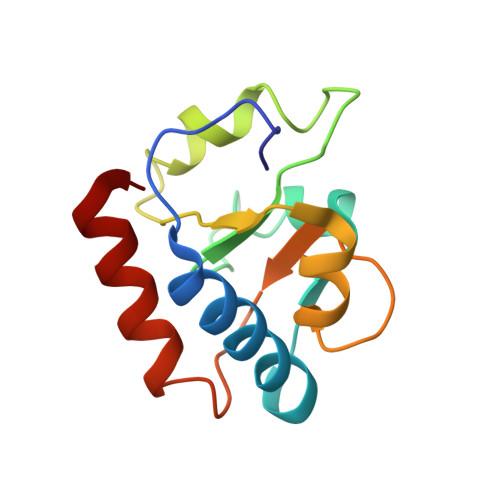
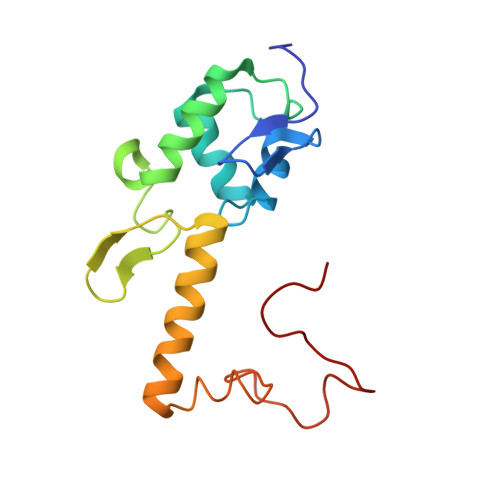
Eukaryotic and archaeal translation initiation complexes have a common structural core comprising e/aIF1, e/aIF1A, the ternary complex (TC, e/aIF2-GTP-Met-tRNA i Met ) and mRNA bound to the small ribosomal subunit. e/aIF2 plays a crucial role in this process but how this factor controls start codon selection remains unclear. Here, we present cryo-EM structures of the full archaeal 30S initiation complex showing two conformational states of the TC. In the first state, the TC is bound to the ribosome in a relaxed conformation with the tRNA oriented out of the P site. In the second state, the tRNA is accommodated within the peptidyl (P) site and the TC becomes constrained. This constraint is compensated by codon/anticodon base pairing, whereas in the absence of a start codon, aIF2 contributes to swing out the tRNA. This spring force concept highlights a mechanism of codon/anticodon probing by the initiator tRNA directly assisted by aIF2.
- Laboratoire de Biochimie, Ecole polytechnique, CNRS, Université Paris-Saclay, 91128 Palaiseau cedex, France.
Organizational Affiliation: