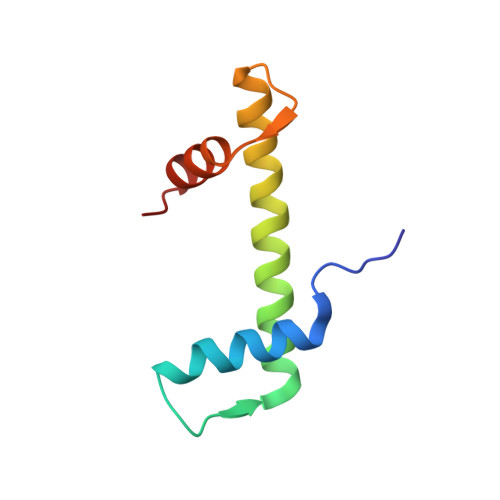
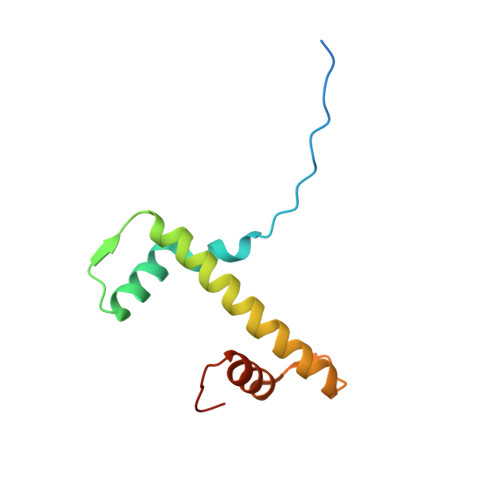
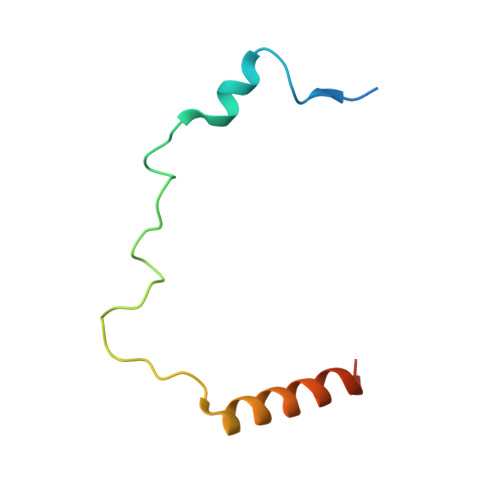
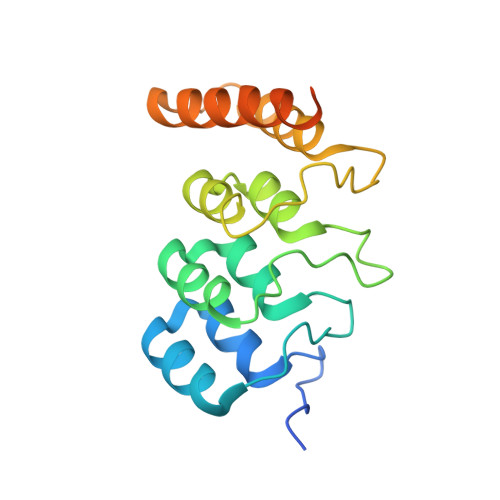
H4K20me0 marks post-replicative chromatin and recruits the TONSL-MMS22L DNA repair complex.
Saredi, G., Huang, H., Hammond, C.M., Alabert, C., Bekker-Jensen, S., Forne, I., Reveron-Gomez, N., Foster, B.M., Mlejnkova, L., Bartke, T., Cejka, P., Mailand, N., Imhof, A., Patel, D.J., Groth, A.(2016) Nature 534: 714-718
- PubMed: 27338793
- DOI: https://doi.org/10.1038/nature18312
- Primary Citation of Related Structures:
5JA4 - PubMed Abstract:
After DNA replication, chromosomal processes including DNA repair and transcription take place in the context of sister chromatids. While cell cycle regulation can guide these processes globally, mechanisms to distinguish pre- and post-replicative states locally remain unknown. Here we reveal that new histones incorporated during DNA replication provide a signature of post-replicative chromatin, read by the human TONSL–MMS22L homologous recombination complex. We identify the TONSL ankyrin repeat domain (ARD) as a reader of histone H4 tails unmethylated at K20 (H4K20me0), which are specific to new histones incorporated during DNA replication and mark post-replicative chromatin until the G2/M phase of the cell cycle. Accordingly, TONSL–MMS22L binds new histones H3–H4 both before and after incorporation into nucleosomes, remaining on replicated chromatin until late G2/M. H4K20me0 recognition is required for TONSL–MMS22L binding to chromatin and accumulation at challenged replication forks and DNA lesions. Consequently, TONSL ARD mutants are toxic, compromising genome stability, cell viability and resistance to replication stress. Together, these data reveal a histone-reader-based mechanism for recognizing the post-replicative state, offering a new angle to understand DNA repair with the potential for targeted cancer therapy.
- Biotech Research and Innovation Centre (BRIC), Faculty of Health and Medical Sciences, University of Copenhagen, Denmark.
Organizational Affiliation: