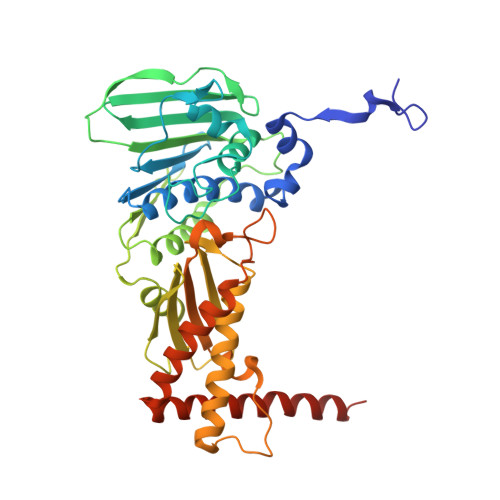
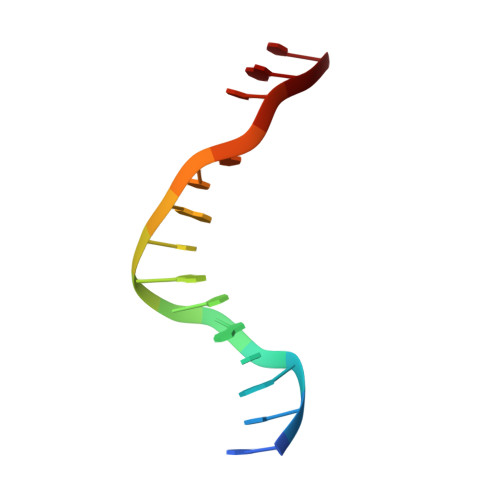
Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase.
Laponogov, I., Pan, X.S., Veselkov, D.A., Skamrova, G.B., Umrekar, T.R., Fisher, L.M., Sanderson, M.R.(2018) Nat Commun 9: 2579-2579
- PubMed: 29968711
- DOI: https://doi.org/10.1038/s41467-018-05005-x
- Primary Citation of Related Structures:
5J5P, 5J5Q - PubMed Abstract:
Type II topoisomerases alter DNA topology to control DNA supercoiling and chromosome segregation and are targets of clinically important anti-infective and anticancer therapeutics. They act as ATP-operated clamps to trap a DNA helix and transport it through a transient break in a second DNA. Here, we present the first X-ray crystal structure solved at 2.83 Å of a closed clamp complete with trapped T-segment DNA obtained by co-crystallizing the ATPase domain of S. pneumoniae topoisomerase IV with a nonhydrolyzable ATP analogue and 14-mer duplex DNA. The ATPase dimer forms a 22 Å protein hole occupied by the kinked DNA bound asymmetrically through positively charged residues lining the hole, and whose mutagenesis impacts the DNA decatenation, DNA relaxation and DNA-dependent ATPase activities of topo IV. These results and a side-bound DNA-ParE structure help explain how the T-segment DNA is captured and transported by a type II topoisomerase, and reveal a new enzyme-DNA interface for drug discovery.
- Randall Centre for Cell and Molecular Biophysics, 3rd Floor New Hunt's House, Faculty of Life Sciences and Medicine, King's College London, London, SE1 1UL, UK.
Organizational Affiliation: