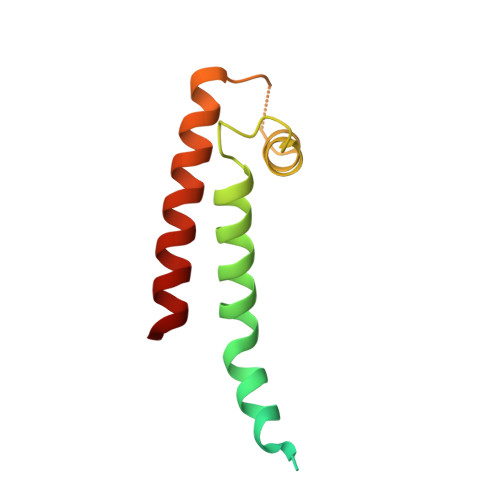
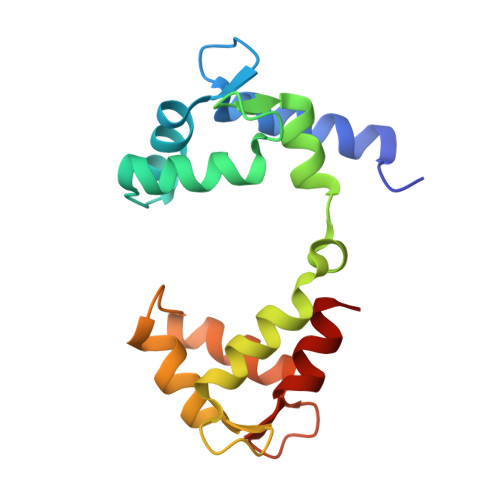
Structural Insights into the M-Channel Proximal C-Terminus/Calmodulin Complex.
Strulovich, R., Tobelaim, W.S., Attali, B., Hirsch, J.A.(2016) Biochemistry 55: 5353-5365
- PubMed: 27564677
- DOI: https://doi.org/10.1021/acs.biochem.6b00477
- Primary Citation of Related Structures:
5J03 - PubMed Abstract:
The Kv7 (KCNQ) channel family, comprising voltage-gated potassium channels, plays major roles in fine-tuning cellular excitability by reducing firing frequency and controlling repolarization. Kv7 channels have a unique intracellular C-terminal (CT) domain bound constitutively by calmodulin (CaM). This domain plays key functions in channel tetramerization, trafficking, and gating. CaM binds to the proximal CT, comprising helices A and B. Kv7.2 and Kv7.3 are expressed in neural tissues. Together, they form the heterotetrameric M channel. We characterized Kv7.2, Kv7.3, and chimeric Kv7.3 helix A-Kv7.2 helix B (Q3A-Q2B) proximal CT/CaM complexes by solution methods at various Ca(2+)concentrations and determined them all to have a 1:1 stoichiometry. We then determined the crystal structure of the Q3A-Q2B/CaM complex at high Ca(2+) concentration to 2.0 Å resolution. CaM hugs the antiparallel coiled coil of helices A and B, braced together by an additional helix. The structure displays a hybrid apo-Ca(2+) CaM conformation even though four Ca(2+) ions are bound. Our results pinpoint unique interactions enabling the possible intersubunit pairing of Kv7.3 helix A and Kv7.2 helix B while underlining the potential importance of Kv7.3 helix A's role in stabilizing channel oligomerization. Also, the structure can be used to rationalize various channelopathic mutants. Functional testing of the chimeric channel found it to have a voltage-dependence similar to the M channel, thereby demonstrating helix A's importance in imparting gating properties.
- Department of Biochemistry and Molecular Biology, Institute of Structural Biology, George S. Wise Faculty of Life Sciences, ‡Department of Physiology and Pharmacology, Sackler Faculty of Medicine, and §Sagol School of Neuroscience, Tel Aviv University , Ramat Aviv 69978, Israel.
Organizational Affiliation: