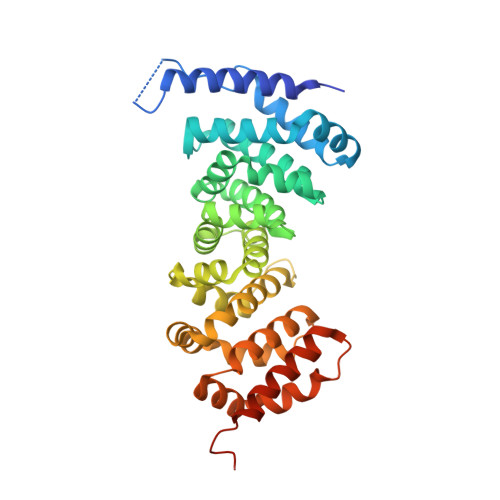
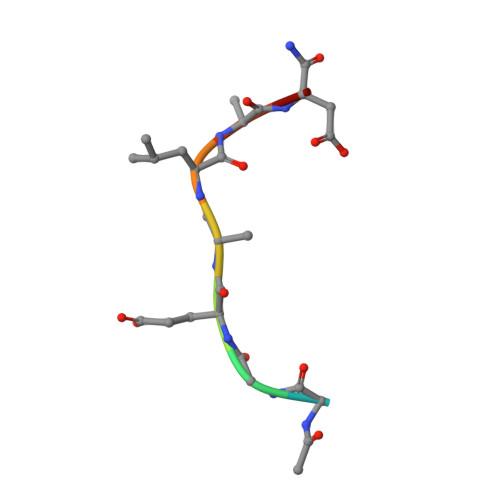
Peptidomimetic inhibitors of APC-Asef interaction block colorectal cancer migration.
Jiang, H., Deng, R., Yang, X., Shang, J., Lu, S., Zhao, Y., Song, K., Liu, X., Zhang, Q., Chen, Y., Chinn, Y.E., Wu, G., Li, J., Chen, G., Yu, J., Zhang, J.(2017) Nat Chem Biol 13: 994-1001
- PubMed: 28759015
- DOI: https://doi.org/10.1038/nchembio.2442
- Primary Citation of Related Structures:
5B6G, 5IZ6, 5IZ8, 5IZ9, 5IZA - PubMed Abstract:
The binding of adenomatous polyposis coli (APC) to its receptor Asef relieves the negative intramolecular regulation of Asef and leads to aberrant cell migration in human colorectal cancer. Because of its crucial role in metastatic dissemination, the interaction between APC and Asef is an attractive target for anti-colorectal-cancer therapy. We rationally designed a series of peptidomimetics that act as potent inhibitors of the APC interface. Crystal structures and biochemical and cellular assays showed that the peptidomimetics in the APC pocket inhibited the migration of colorectal cells by disrupting APC-Asef interaction. By using the peptidomimetic inhibitor as a chemical probe, we found that CDC42 was the downstream GTPase involved in APC-stimulated Asef activation in colorectal cancer cells. Our work demonstrates the feasibility of exploiting APC-Asef interaction to regulate the migration of colorectal cancer cells, and provides what to our knowledge is the first class of protein-protein interaction inhibitors available for the development of cancer therapeutics targeting APC-Asef signaling.
- Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Ministry of Education, Shanghai Jiao-Tong University School of Medicine, Shanghai, China.
Organizational Affiliation: