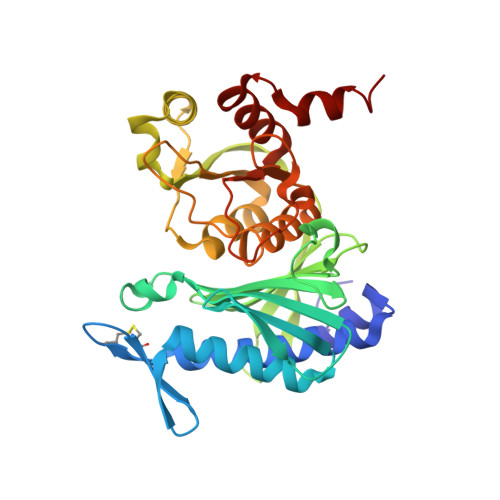
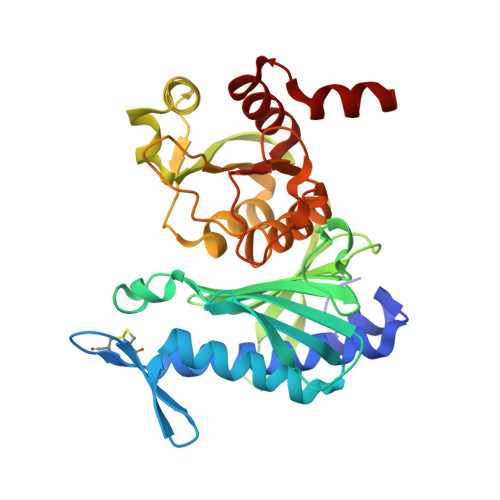
Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories.
Gutle, D.D., Roret, T., Muller, S.J., Couturier, J., Lemaire, S.D., Hecker, A., Dhalleine, T., Buchanan, B.B., Reski, R., Einsle, O., Jacquot, J.P.(2016) Proc Natl Acad Sci U S A 113: 6779-6784
- PubMed: 27226308
- DOI: https://doi.org/10.1073/pnas.1606241113
- Primary Citation of Related Structures:
5IZ1, 5IZ3 - PubMed Abstract:
The Calvin-Benson cycle of carbon dioxide fixation in chloroplasts is controlled by light-dependent redox reactions that target specific enzymes. Of the regulatory members of the cycle, our knowledge of sedoheptulose-1,7-bisphosphatase (SBPase) is particularly scanty, despite growing evidence for its importance and link to plant productivity. To help fill this gap, we have purified, crystallized, and characterized the recombinant form of the enzyme together with the better studied fructose-1,6-bisphosphatase (FBPase), in both cases from the moss Physcomitrella patens (Pp). Overall, the moss enzymes resembled their counterparts from seed plants, including oligomeric organization-PpSBPase is a dimer, and PpFBPase is a tetramer. The two phosphatases showed striking structural homology to each other, differing primarily in their solvent-exposed surface areas in a manner accounting for their specificity for seven-carbon (sedoheptulose) and six-carbon (fructose) sugar bisphosphate substrates. The two enzymes had a similar redox potential for their regulatory redox-active disulfides (-310 mV for PpSBPase vs. -290 mV for PpFBPase), requirement for Mg(2+) and thioredoxin (TRX) specificity (TRX f > TRX m). Previously known to differ in the position and sequence of their regulatory cysteines, the enzymes unexpectedly showed unique evolutionary histories. The FBPase gene originated in bacteria in conjunction with the endosymbiotic event giving rise to mitochondria, whereas SBPase arose from an archaeal gene resident in the eukaryotic host. These findings raise the question of how enzymes with such different evolutionary origins achieved structural similarity and adapted to control by the same light-dependent photosynthetic mechanism-namely ferredoxin, ferredoxin-thioredoxin reductase, and thioredoxin.
- Plant Biotechnology, Faculty of Biology, University of Freiburg, 79104 Freiburg, Germany; Université de Lorraine, UMR 1136 Interactions Arbres Microorganismes, F-54500 Vandœuvre-les-Nancy, France; Institut national de la recherche agronomique (INRA), UMR 1136 Interactions Arbres Microorganismes, F-54280 Champenoux, France; Spemann Graduate School of Biology and Medicine, University of Freiburg, 79104 Freiburg, Germany;
Organizational Affiliation: