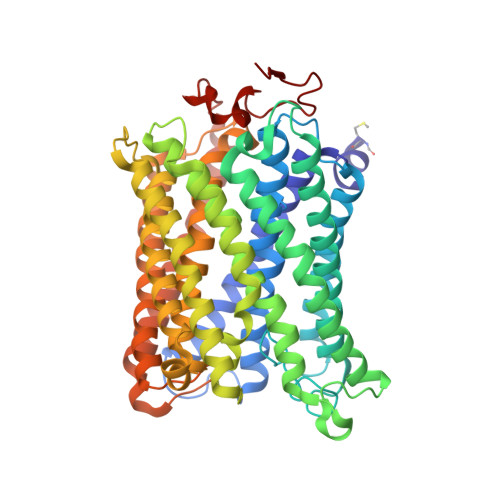
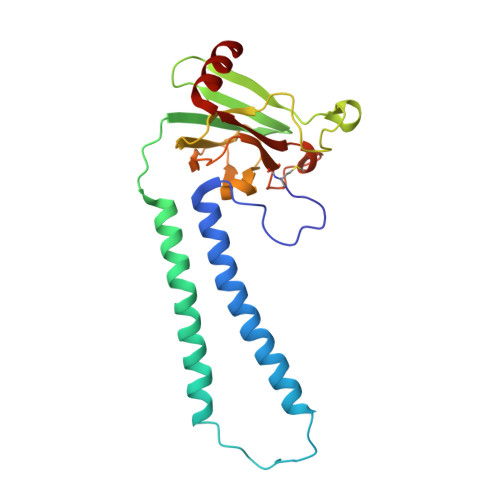
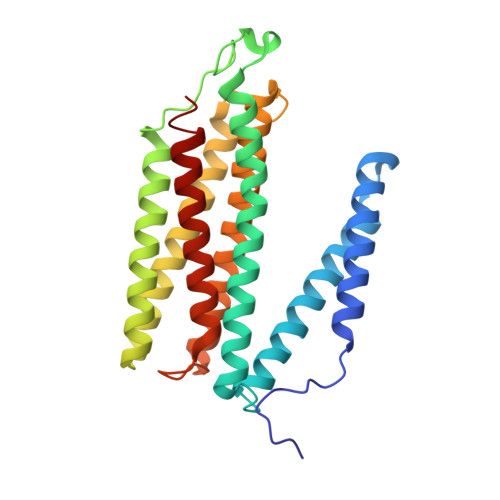
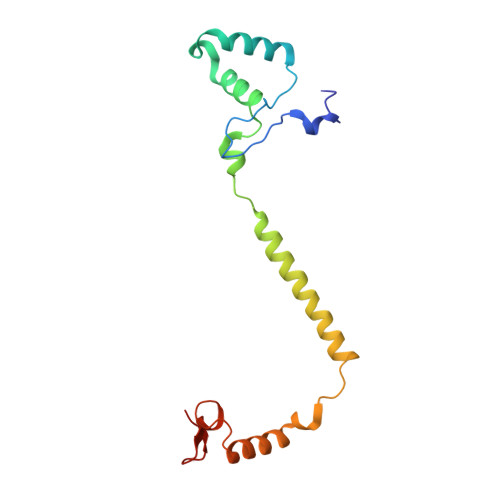
Complex structure of cytochrome c-cytochrome c oxidase reveals a novel protein-protein interaction mode
Shimada, S., Shinzawa-Itoh, K., Baba, J., Aoe, S., Shimada, A., Yamashita, E., Kang, J., Tateno, M., Yoshikawa, S., Tsukihara, T.(2017) EMBO J 36: 291-300
- PubMed: 27979921
- DOI: https://doi.org/10.15252/embj.201695021
- Primary Citation of Related Structures:
5IY5 - PubMed Abstract:
Mitochondrial cytochrome c oxidase (CcO) transfers electrons from cytochrome c (Cyt.c) to O 2 to generate H 2 O, a process coupled to proton pumping. To elucidate the mechanism of electron transfer, we determined the structure of the mammalian Cyt.c-CcO complex at 2.0-Å resolution and identified an electron transfer pathway from Cyt.c to CcO. The specific interaction between Cyt.c and CcO is stabilized by a few electrostatic interactions between side chains within a small contact surface area. Between the two proteins are three water layers with a long inter-molecular span, one of which lies between the other two layers without significant direct interaction with either protein. Cyt.c undergoes large structural fluctuations, using the interacting regions with CcO as a fulcrum. These features of the protein-protein interaction at the docking interface represent the first known example of a new class of protein-protein interaction, which we term "soft and specific". This interaction is likely to contribute to the rapid association/dissociation of the Cyt.c-CcO complex, which facilitates the sequential supply of four electrons for the O 2 reduction reaction.
- Picobiology Institute, Graduate School of Life Science, University of Hyogo, Akoh, Hyogo, Japan.
Organizational Affiliation: