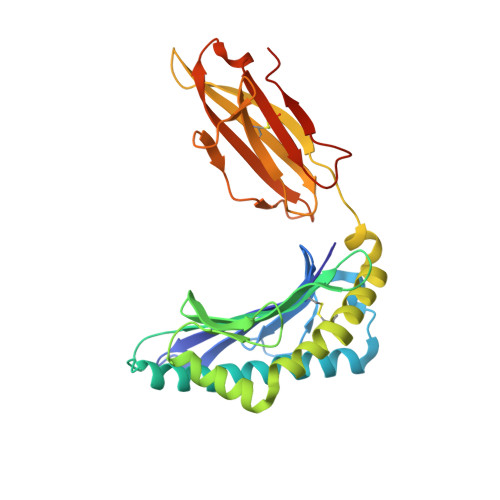
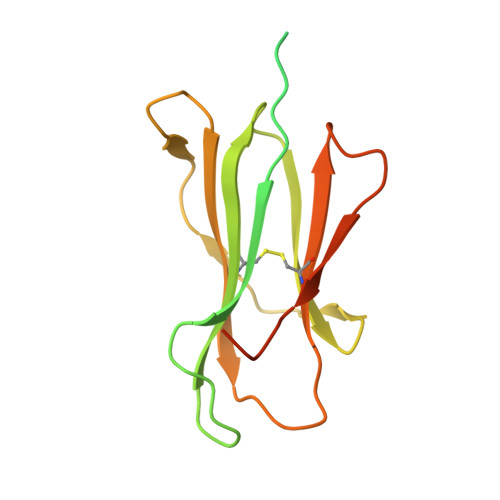
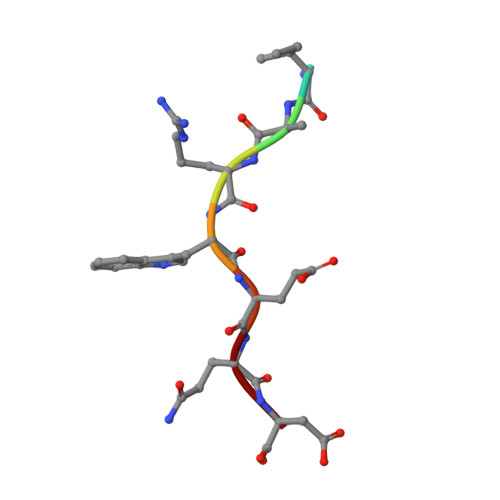
Human Leukocyte Antigen F Presents Peptides and Regulates Immunity through Interactions with NK Cell Receptors.
Dulberger, C.L., McMurtrey, C.P., Holzemer, A., Neu, K.E., Liu, V., Steinbach, A.M., Garcia-Beltran, W.F., Sulak, M., Jabri, B., Lynch, V.J., Altfeld, M., Hildebrand, W.H., Adams, E.J.(2017) Immunity 46: 1018-1029.e7
- PubMed: 28636952
- DOI: https://doi.org/10.1016/j.immuni.2017.06.002
- Primary Citation of Related Structures:
5IUE, 5KNM - PubMed Abstract:
Evidence is mounting that the major histocompatibility complex (MHC) molecule HLA-F (human leukocyte antigen F) regulates the immune system in pregnancy, infection, and autoimmunity by signaling through NK cell receptors (NKRs). We present structural, biochemical, and evolutionary analyses demonstrating that HLA-F presents peptides of unconventional length dictated by a newly arisen mutation (R62W) that has produced an open-ended groove accommodating particularly long peptides. Compared to empty HLA-F open conformers (OCs), HLA-F tetramers bound with human-derived peptides differentially stained leukocytes, suggesting peptide-dependent engagement. Our in vitro studies confirm that NKRs differentiate between peptide-bound and peptide-free HLA-F. The complex structure of peptide-loaded β 2 m-HLA-F bound to the inhibitory LIR1 revealed similarities to high-affinity recognition of the viral MHC-I mimic UL18 and a docking strategy that relies on contacts with HLA-F as well as β 2 m, thus precluding binding to HLA-F OCs. These findings provide a biochemical framework to understand how HLA-F could regulate immunity via interactions with NKRs.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: