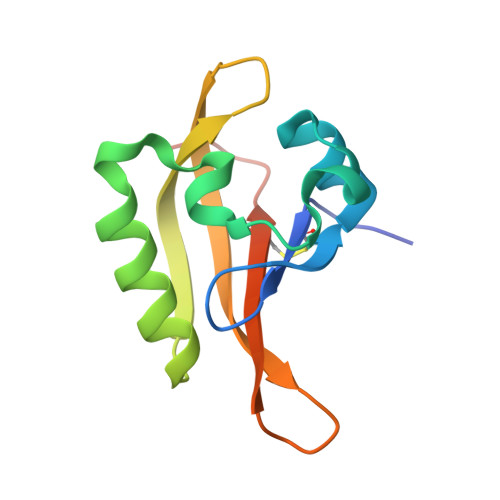
Genetic Analysis of Physcomitrella patens Identifies ABSCISIC ACID NON-RESPONSIVE, a Regulator of ABA Responses Unique to Basal Land Plants and Required for Desiccation Tolerance.
Stevenson, S.R., Kamisugi, Y., Trinh, C.H., Schmutz, J., Jenkins, J.W., Grimwood, J., Muchero, W., Tuskan, G.A., Rensing, S.A., Lang, D., Reski, R., Melkonian, M., Rothfels, C.J., Li, F.W., Larsson, A., Wong, G.K., Edwards, T.A., Cuming, A.C.(2016) Plant Cell 28: 1310-1327
- PubMed: 27194706
- DOI: https://doi.org/10.1105/tpc.16.00091
- Primary Citation of Related Structures:
5IU1 - PubMed Abstract:
The anatomically simple plants that first colonized land must have acquired molecular and biochemical adaptations to drought stress. Abscisic acid (ABA) coordinates responses leading to desiccation tolerance in all land plants. We identified ABA nonresponsive mutants in the model bryophyte Physcomitrella patens and genotyped a segregating population to map and identify the ABA NON-RESPONSIVE (ANR) gene encoding a modular protein kinase comprising an N-terminal PAS domain, a central EDR domain, and a C-terminal MAPKKK-like domain. anr mutants fail to accumulate dehydration tolerance-associated gene products in response to drought, ABA, or osmotic stress and do not acquire ABA-dependent desiccation tolerance. The crystal structure of the PAS domain, determined to 1.7-Å resolution, shows a conserved PAS-fold that dimerizes through a weak dimerization interface. Targeted mutagenesis of a conserved tryptophan residue within the PAS domain generates plants with ABA nonresponsive growth and strongly attenuated ABA-responsive gene expression, whereas deleting this domain retains a fully ABA-responsive phenotype. ANR orthologs are found in early-diverging land plant lineages and aquatic algae but are absent from more recently diverged vascular plants. We propose that ANR genes represent an ancestral adaptation that enabled drought stress survival of the first terrestrial colonizers but were lost during land plant evolution.
- Centre for Plant Sciences, Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, United Kingdom.
Organizational Affiliation: