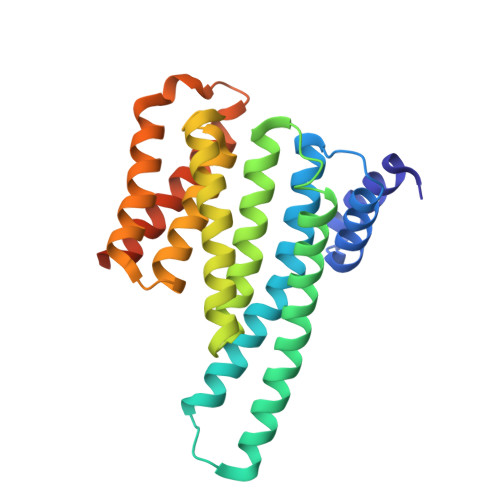
Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways
Xiao, B., Smerdon, S.J., Jones, D.H., Dodson, G.G., Soneji, Y., Aitken, A., Gamblin, S.J.(1995) Nature 376: 188-191
- PubMed: 7603573
- DOI: https://doi.org/10.1038/376188a0
- Primary Citation of Related Structures:
5IQP - PubMed Abstract:
A broad range of organisms and tissues contain 14-3-3 proteins, which have been associated with many diverse functions including critical roles in signal transduction pathways, exocytosis and cell cycle regulation. We report here the crystal structure of the human T-cell 14-3-3 isoform (tau) dimer at 2.6 A resolution. Each monomer (Mr 28K) is composed of an unusual arrangement of nine antiparallel alpha-helices organized as two structural domains. The dimer creates a large, negatively charged channel approximately 35 A broad, 35 A wide and 20 A deep. Overall, invariant residues line the interior of this channel whereas the more variable residues are distributed on the outer surface. At the base of this channel is a 16-residue segment of 14-3-3 which has been implicated in the binding of 14-3-3 to protein kinase C.
- Division of Protein Structure, National Institute for Medical Research, Mill Hill, London, UK.
Organizational Affiliation: