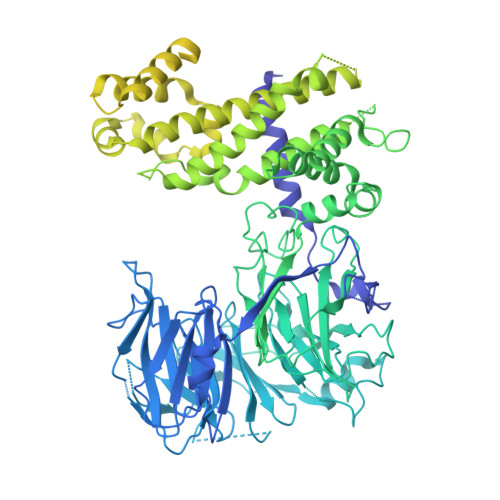
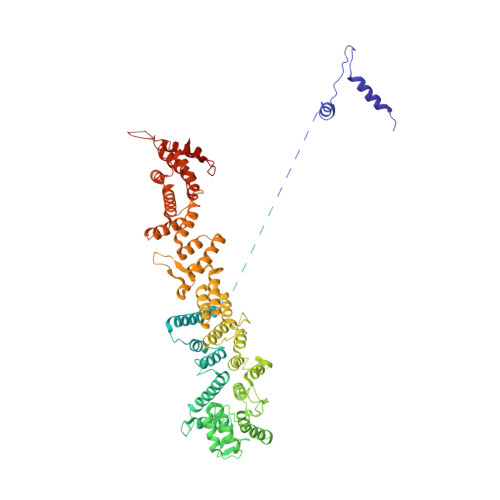
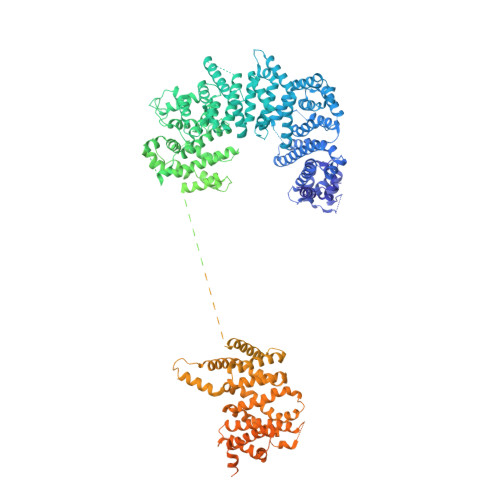
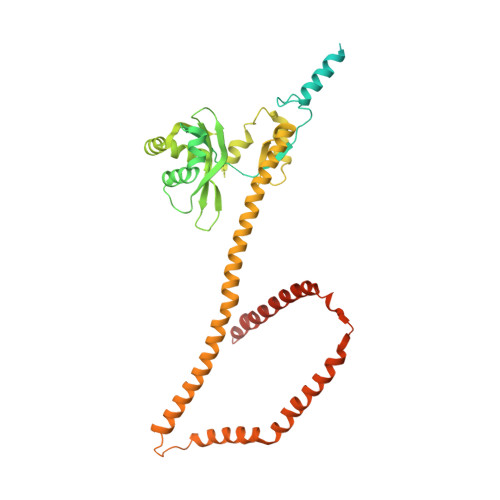
Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Kosinski, J., Mosalaganti, S., von Appen, A., Teimer, R., DiGuilio, A.L., Wan, W., Bui, K.H., Hagen, W.J., Briggs, J.A., Glavy, J.S., Hurt, E., Beck, M.(2016) Science 352: 363-365
- PubMed: 27081072
- DOI: https://doi.org/10.1126/science.aaf0643
- Primary Citation of Related Structures:
5IJN, 5IJO - PubMed Abstract:
Nuclear pore complexes (NPCs) are 110-megadalton assemblies that mediate nucleocytoplasmic transport. NPCs are built from multiple copies of ~30 different nucleoporins, and understanding how these nucleoporins assemble into the NPC scaffold imposes a formidable challenge. Recently, it has been shown how the Y complex, a prominent NPC module, forms the outer rings of the nuclear pore. However, the organization of the inner ring has remained unknown until now. We used molecular modeling combined with cross-linking mass spectrometry and cryo-electron tomography to obtain a composite structure of the inner ring. This architectural map explains the vast majority of the electron density of the scaffold. We conclude that despite obvious differences in morphology and composition, the higher-order structure of the inner and outer rings is unexpectedly similar.
- Structural and Computational Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany.
Organizational Affiliation: