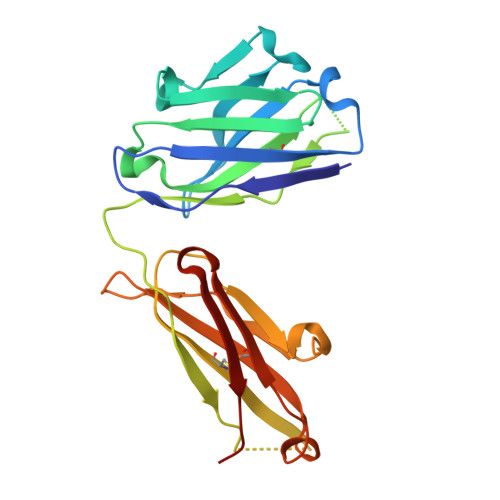
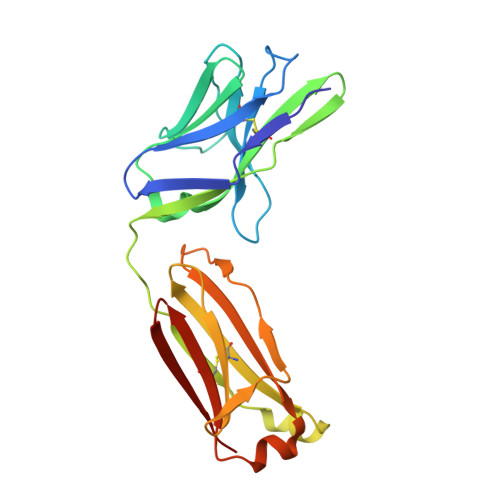
Stereotyped antibody responses target posttranslationally modified gluten in celiac disease.
Snir, O., Chen, X., Gidoni, M., du Pre, M.F., Zhao, Y., Steinsbo, O., Lundin, K.E., Yaari, G., Sollid, L.M.(2017) JCI Insight 2
- PubMed: 28878138
- DOI: https://doi.org/10.1172/jci.insight.93961
- Primary Citation of Related Structures:
5IFJ, 5IG7, 5IHZ, 5IJK, 5IK3 - PubMed Abstract:
The role of B cells and posttranslational modifications in pathogenesis of organ-specific immune diseases is increasingly envisioned but remains poorly understood, particularly in human disorders. In celiac disease, transglutaminase 2-modified (TG2-modified; deamidated) gluten peptides drive disease-specific T cell and B cell responses, and antibodies to deamidated gluten peptides are excellent diagnostic markers. Here, we substantiate by high-throughput sequencing of IGHV genes that antibodies to a disease-specific, deamidated, and immunodominant B cell epitope of gluten (PLQPEQPFP) have biased and stereotyped usage of IGHV3-23 and IGHV3-15 gene segments with modest somatic mutations. X-ray crystal structures of 2 prototype IGHV3-15/IGKV4-1 and IGHV3-23/IGLV4-69 antibodies reveal peptide interaction mainly via germline-encoded residues. In-depth mutational analysis showed restricted selection and substitution patterns at positions involved in antigen binding. While the IGHV3-15/IGKV4-1 antibody interacts with Glu5 and Gln6, the IGHV3-23/IGLV4-69 antibody interacts with Gln3, Pro4, Pro7, and Phe8 - residues involved in substrate recognition by TG2. Hence, both antibodies, despite different interaction with the epitope, recognize signatures of TG2 processing that facilitates B cell presentation of deamidated gluten peptides to T cells, thereby providing a molecular framework for the generation of these clinically important antibodies. The study provides essential insight into the pathogenic mechanism of celiac disease.
- Centre for Immune Regulation and Department of Immunology, University of Oslo and Oslo University Hospital-Rikshospitalet, Oslo, Norway.
Organizational Affiliation: