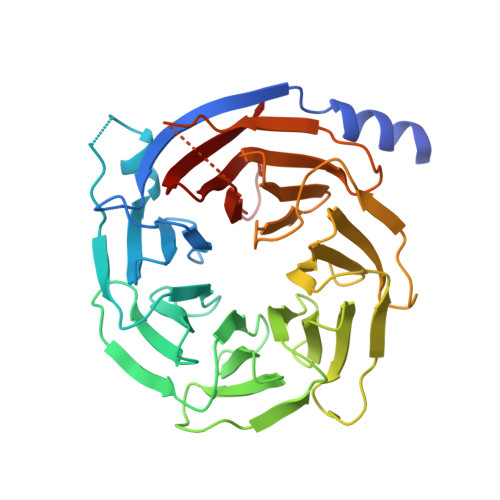
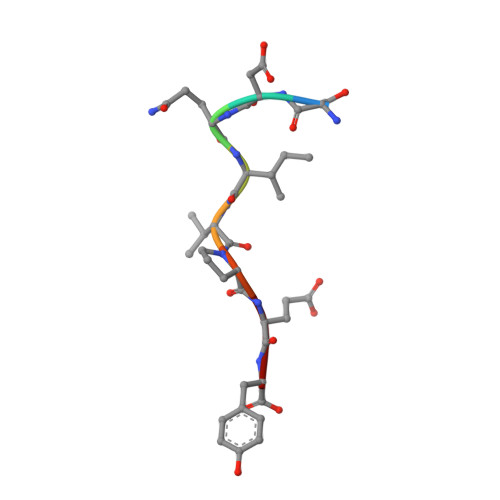
Structural Basis for Substrate Selectivity of the E3 Ligase COP1.
Uljon, S., Xu, X., Durzynska, I., Stein, S., Adelmant, G., Marto, J.A., Pear, W.S., Blacklow, S.C.(2016) Structure 24: 687-696
- PubMed: 27041596
- DOI: https://doi.org/10.1016/j.str.2016.03.002
- Primary Citation of Related Structures:
5HQG, 5IGO, 5IGQ - PubMed Abstract:
COP1 proteins are E3 ubiquitin ligases that regulate phototropism in plants and target transcription factors for degradation in mammals. The substrate-binding region of COP1 resides within a WD40-repeat domain that also binds to Trib proteins, which are adaptors for C/EBPα degradation. Here we report structures of the human COP1 WD40 domain in isolation, and complexes of the human and Arabidopsis thaliana COP1 WD40 domains with the binding motif of Trib1. The human and Arabidopsis WD40 domains are seven-bladed β propellers with an inserted loop on the bottom face of the first blade. The Trib1 peptide binds in an extended conformation to a highly conserved surface on the top face of the β propeller, indicating a general mode for recognition of peptide motifs by COP1. Together, these studies identify the structural basis and key interactions for motif recognition by COP1, and hint at how Trib1 autoinhibition is overcome to target C/EBPα for degradation.
- Department of Cancer Biology, Dana Farber Cancer Institute, Boston, MA 02215, USA; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA; Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: