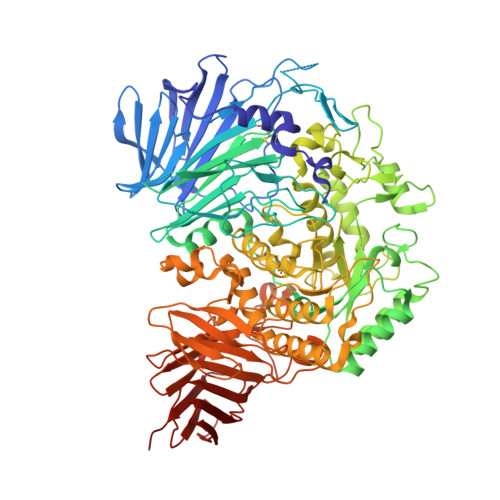
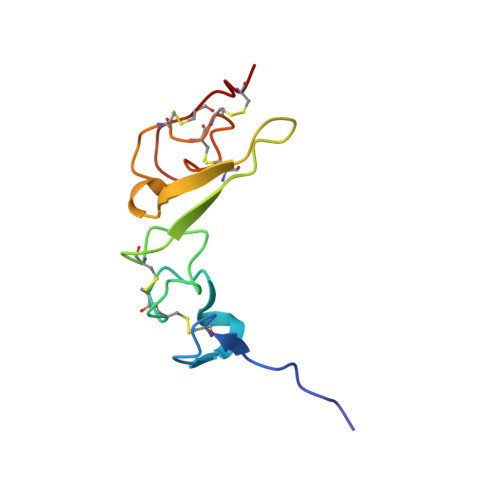
Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Caputo, A.T., Alonzi, D.S., Marti, L., Reca, I.B., Kiappes, J.L., Struwe, W.B., Cross, A., Basu, S., Lowe, E.D., Darlot, B., Santino, A., Roversi, P., Zitzmann, N.(2016) Proc Natl Acad Sci U S A 113: E4630-E4638
- PubMed: 27462106
- DOI: https://doi.org/10.1073/pnas.1604463113
- Primary Citation of Related Structures:
5F0E, 5H9O, 5HJO, 5HJR, 5IED, 5IEE, 5IEF, 5IEG - PubMed Abstract:
The biosynthesis of enveloped viruses depends heavily on the host cell endoplasmic reticulum (ER) glycoprotein quality control (QC) machinery. This dependency exceeds the dependency of host glycoproteins, offering a window for the targeting of ERQC for the development of broad-spectrum antivirals. We determined small-angle X-ray scattering (SAXS) and crystal structures of the main ERQC enzyme, ER α-glucosidase II (α-GluII; from mouse), alone and in complex with key ligands of its catalytic cycle and antiviral iminosugars, including two that are in clinical trials for the treatment of dengue fever. The SAXS data capture the enzyme's quaternary structure and suggest a conformational rearrangement is needed for the simultaneous binding of a monoglucosylated glycan to both subunits. The X-ray structures with key catalytic cycle intermediates highlight that an insertion between the +1 and +2 subsites contributes to the enzyme's activity and substrate specificity, and reveal that the presence of d-mannose at the +1 subsite renders the acid catalyst less efficient during the cleavage of the monoglucosylated substrate. The complexes with iminosugar antivirals suggest that inhibitors targeting a conserved ring of aromatic residues between the α-GluII +1 and +2 subsites would have increased potency and selectivity, thus providing a template for further rational drug design.
- Oxford Glycobiology Institute, Department of Biochemistry, University of Oxford, Oxford OX1 3QU, United Kingdom;
Organizational Affiliation: