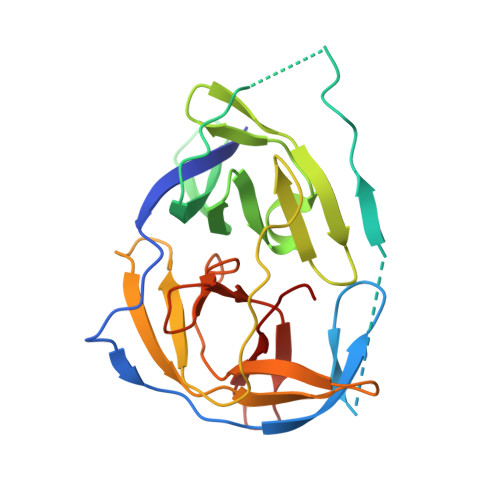
Peptide-Boronic Acid Inhibitors of Flaviviral Proteases: Medicinal Chemistry and Structural Biology.
Nitsche, C., Zhang, L., Weigel, L.F., Schilz, J., Graf, D., Bartenschlager, R., Hilgenfeld, R., Klein, C.D.(2017) J Med Chem 60: 511-516
- PubMed: 27966962
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01021
- Primary Citation of Related Structures:
5IDK - PubMed Abstract:
A thousand-fold affinity gain is achieved by introduction of a C-terminal boronic acid moiety into dipeptidic inhibitors of the Zika, West Nile, and dengue virus proteases. The resulting compounds have K i values in the two-digit nanomolar range, are not cytotoxic, and inhibit virus replication. Structure-activity relationships and a high resolution X-ray cocrystal structure with West Nile virus protease provide a basis for the design of optimized covalent-reversible inhibitors aimed at emerging flaviviral pathogens.
- Medicinal Chemistry, IPMB, Heidelberg University , INF-364, 69120 Heidelberg, Germany.
Organizational Affiliation: