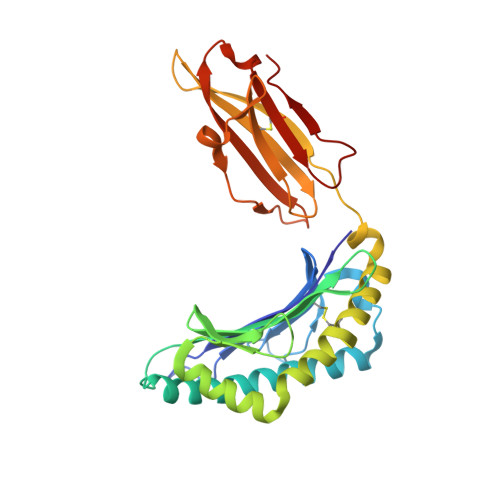
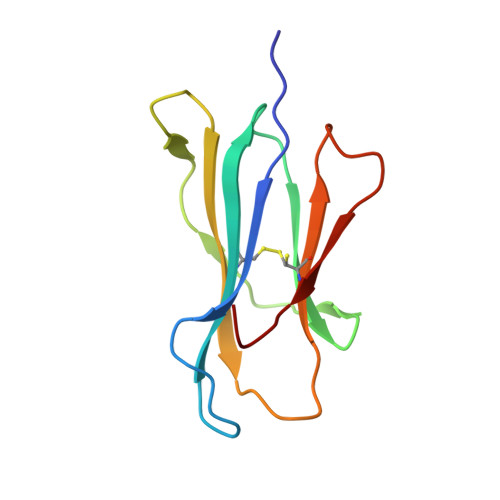
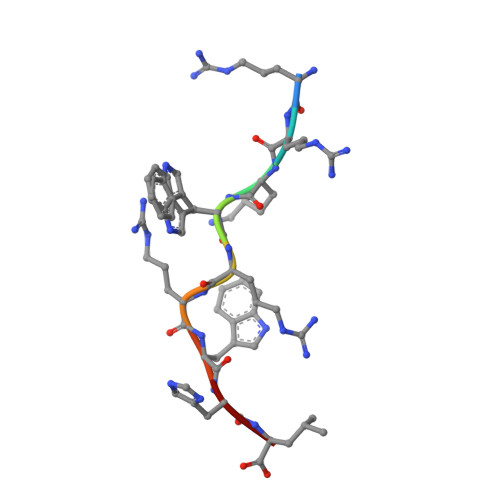
Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
Driller, R., Ballaschk, M., Schmieder, P., Uchanska-Ziegler, B., Ziegler, A., Loll, B.(2019) J Biological Chem
- PubMed: 31296658
- DOI: https://doi.org/10.1074/jbc.RA119.008937
- Primary Citation of Related Structures:
5IB1, 5IB2, 5IB3, 5IB4, 5IB5 - PubMed Abstract:
Conformational changes of major histocompatibility complex (MHC) antigens have the potential to be recognized by T cells and may arise from polymorphic variation of the MHC molecule, the binding of modifying ligands, or both. Here, we investigated whether metal ions could affect allele-dependent structural variation of the two minimally distinct human leukocyte antigen (HLA)-B*27:05 and HLA-B*27:09 subtypes, which exhibit differential association with the rheumatic disease ankylosing spondylitis (AS). We employed NMR spectroscopy and X-ray crystallography coupled with ensemble refinement to study the AS-associated HLA-B*27:05 subtype and the AS-nonassociated HLA-B* 27:09 in complex with the self-peptide pVIPR (RRKWRRWHL). Both techniques revealed that pVIPR exhibits a higher degree of flexibility when complexed with HLA-B*27:05 than with HLA-B*27:09. Furthermore, we found that the binding of the metal ion Cu 2+ or Ni 2+ , but not Mn 2+ , Zn 2+ , or Hg 2+ , affects the structure of a pVIPR-bound HLA-B*27 molecule in a subtype-dependent manner. In HLA-B*27:05, the metals triggered conformational reorientations of pVIPR, but no such structural changes were observed in the HLA-B*27:09 subtype, with or without bound metal ion. These observations provide the first demonstration that not only major histocompatibility complex class II, but also class I, molecules can undergo metal ion-induced conformational alterations. Our findings suggest that metals may have a role in triggering rheumatic diseases such as AS and also have implications for the molecular basis of metal-induced hypersensitivities and allergies.
- Institut für Chemie/Biochemie, AG Strukturbiochemie, Freie Universität Berlin, Takustrasse 6, 14195 Berlin, Germany.
Organizational Affiliation: