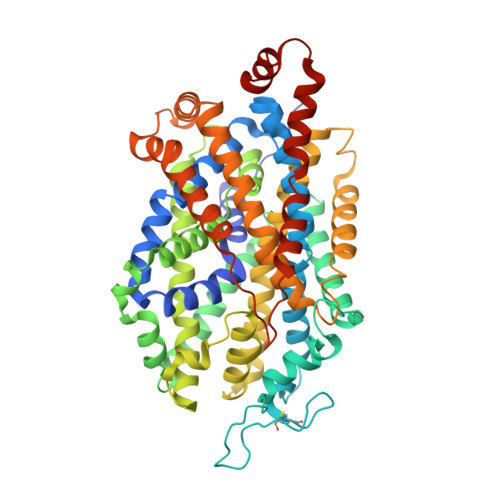
X-ray structures and mechanism of the human serotonin transporter.
Coleman, J.A., Green, E.M., Gouaux, E.(2016) Nature 532: 334-339
- PubMed: 27049939
- DOI: https://doi.org/10.1038/nature17629
- Primary Citation of Related Structures:
5I66, 5I6X, 5I6Z, 5I71, 5I73, 5I74, 5I75 - PubMed Abstract:
The serotonin transporter (SERT) terminates serotonergic signalling through the sodium- and chloride-dependent reuptake of neurotransmitter into presynaptic neurons. SERT is a target for antidepressant and psychostimulant drugs, which block reuptake and prolong neurotransmitter signalling. Here we report X-ray crystallographic structures of human SERT at 3.15 Å resolution bound to the antidepressants (S)-citalopram or paroxetine. Antidepressants lock SERT in an outward-open conformation by lodging in the central binding site, located between transmembrane helices 1, 3, 6, 8 and 10, directly blocking serotonin binding. We further identify the location of an allosteric site in the complex as residing at the periphery of the extracellular vestibule, interposed between extracellular loops 4 and 6 and transmembrane helices 1, 6, 10 and 11. Occupancy of the allosteric site sterically hinders ligand unbinding from the central site, providing an explanation for the action of (S)-citalopram as an allosteric ligand. These structures define the mechanism of antidepressant action in SERT, and provide blueprints for future drug design.
- Vollum Institute, Oregon Health &Science University, Portland, Oregon 97239, USA.
Organizational Affiliation: