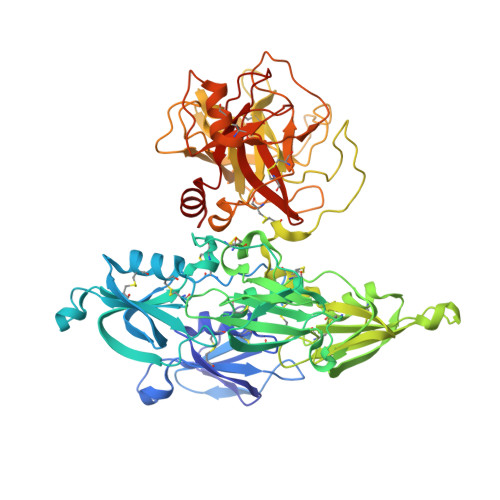
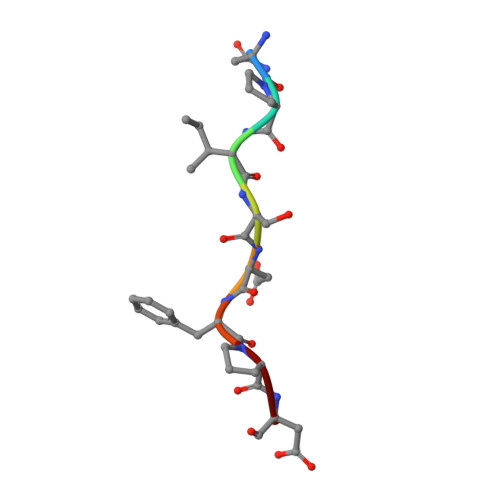
A novel DFP tripeptide motif interacts with the coagulation factor XI apple 2 domain.
Wong, S.S., stergaard, S., Hall, G., Li, C., Williams, P.M., Stennicke, H., Emsley, J.(2016) Blood 127: 2915-2923
- PubMed: 27006387
- DOI: https://doi.org/10.1182/blood-2015-10-676122
- Primary Citation of Related Structures:
5EOD, 5EOK, 5I25 - PubMed Abstract:
Factor XI (FXI) is the zymogen of FXIa, which cleaves FIX in the intrinsic pathway of coagulation. FXI is known to exist as a dimer and interacts with multiple proteins via its 4 apple domains in the "saucer section" of the enzyme; however, to date, no complex crystal structure has been described. To investigate protein interactions of FXI, a large random peptide library consisting of 10(6) to 10(7) peptides was screened for FXI binding, which identified a series of FXI binding motifs containing the signature Asp-Phe-Pro (DFP) tripeptide. Motifs containing this core tripeptide were found in diverse proteins, including the known ligand high-molecular-weight kininogen (HK), as well as the extracellular matrix proteins laminin and collagen V. To define the binding site on FXI, we determined the crystal structure of FXI in complex with the HK-derived peptide NPISDFPDT. This revealed the location of the DFP peptide bound to the FXI apple 2 domain, and central to the interaction, the DFP phenylalanine side-chain inserts into a major hydrophobic pocket in the apple 2 domain and the isoleucine occupies a flanking minor pocket. Two further structures of FXI in complex with the laminin-derived peptide EFPDFP and a DFP peptide from the random screen demonstrated binding in the same pocket, although in a slightly different conformation, thus revealing some flexibility in the molecular interactions of the FXI apple 2 domain.
- Centre for Biomolecular Sciences, School of Pharmacy, University of Nottingham, Nottingham, United Kingdom; and Novo Nordisk, Måløv, Denmark.
Organizational Affiliation: