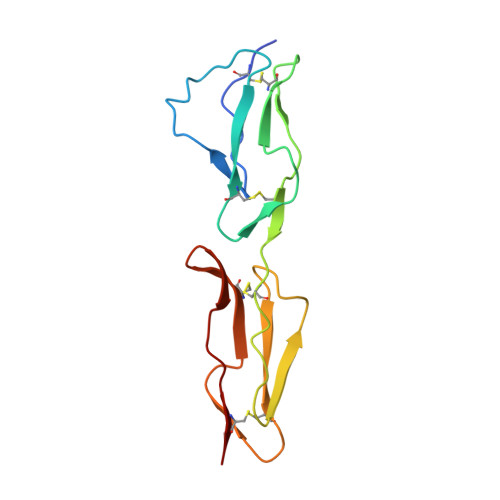
Conserved patterns hidden within group A Streptococcus M protein hypervariability recognize human C4b-binding protein.
Buffalo, C.Z., Bahn-Suh, A.J., Hirakis, S.P., Biswas, T., Amaro, R.E., Nizet, V., Ghosh, P.(2016) Nat Microbiol 1: 16155-16155
- PubMed: 27595425
- DOI: https://doi.org/10.1038/nmicrobiol.2016.155
- Primary Citation of Related Structures:
5HYP, 5HYT, 5HYU, 5HZP, 5I0Q - PubMed Abstract:
No vaccine exists against group A Streptococcus (GAS), a leading cause of worldwide morbidity and mortality. A severe hurdle is the hypervariability of its major antigen, the M protein, with >200 different M types known. Neutralizing antibodies typically recognize M protein hypervariable regions (HVRs) and confer narrow protection. In stark contrast, human C4b-binding protein (C4BP), which is recruited to the GAS surface to block phagocytic killing, interacts with a remarkably large number of M protein HVRs (apparently ∼90%). Such broad recognition is rare, and we discovered a unique mechanism for this through the structure determination of four sequence-diverse M proteins in complexes with C4BP. The structures revealed a uniform and tolerant 'reading head' in C4BP, which detected conserved sequence patterns hidden within hypervariability. Our results open up possibilities for rational therapies that target the M-C4BP interaction, and also inform a path towards vaccine design.
- Department of Chemistry &Biochemistry, University of California, San Diego, La Jolla, California 92093, USA.
Organizational Affiliation: