A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase.
Matyskiela, M.E., Lu, G., Ito, T., Pagarigan, B., Lu, C.C., Miller, K., Fang, W., Wang, N.Y., Nguyen, D., Houston, J., Carmel, G., Tran, T., Riley, M., Nosaka, L., Lander, G.C., Gaidarova, S., Xu, S., Ruchelman, A.L., Handa, H., Carmichael, J., Daniel, T.O., Cathers, B.E., Lopez-Girona, A., Chamberlain, P.P.(2016) Nature 535: 252-257
- PubMed: 27338790
- DOI: https://doi.org/10.1038/nature18611
- Primary Citation of Related Structures:
5HXB - PubMed Abstract:
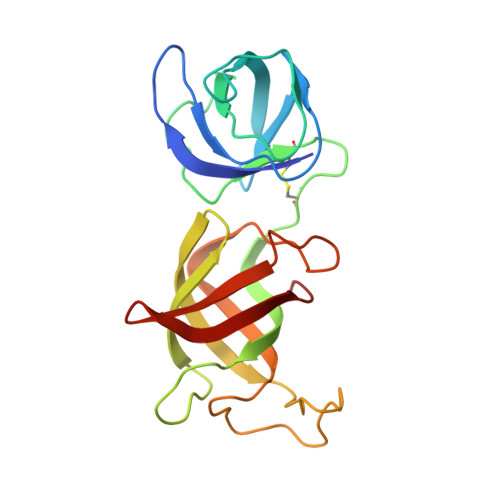
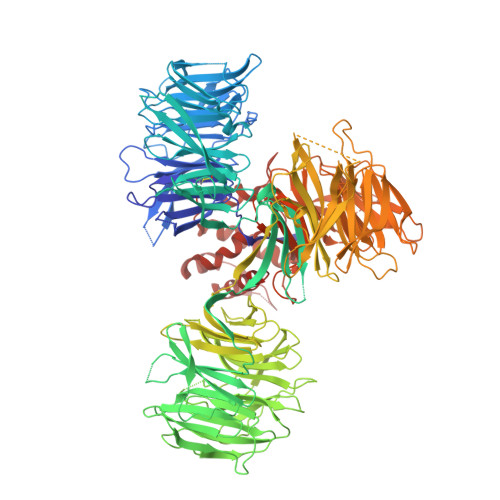
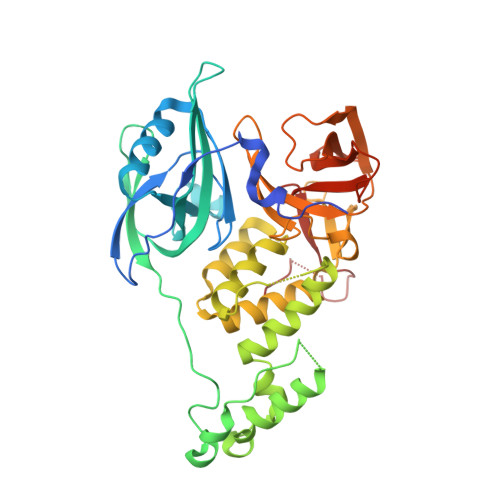
Immunomodulatory drugs bind to cereblon (CRBN) to confer differentiated substrate specificity on the CRL4(CRBN) E3 ubiquitin ligase. Here we report the identification of a new cereblon modulator, CC-885, with potent anti-tumour activity. The anti-tumour activity of CC-885 is mediated through the cereblon-dependent ubiquitination and degradation of the translation termination factor GSPT1. Patient-derived acute myeloid leukaemia tumour cells exhibit high sensitivity to CC-885, indicating the clinical potential of this mechanism. Crystallographic studies of the CRBN-DDB1-CC-885-GSPT1 complex reveal that GSPT1 binds to cereblon through a surface turn containing a glycine residue at a key position, interacting with both CC-885 and a 'hotspot' on the cereblon surface. Although GSPT1 possesses no obvious structural, sequence or functional homology to previously known cereblon substrates, mutational analysis and modelling indicate that the cereblon substrate Ikaros uses a similar structural feature to bind cereblon, suggesting a common motif for substrate recruitment. These findings define a structural degron underlying cereblon 'neosubstrate' selectivity, and identify an anti-tumour target rendered druggable by cereblon modulation.
- Celgene Corporation, 10300 Campus Point Drive, Suite 100, San Diego, California 92121, USA.
Organizational Affiliation: