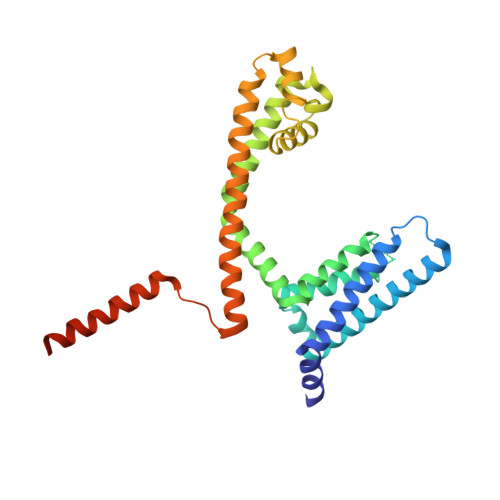
The complete structure of an activated open sodium channel.
Sula, A., Booker, J., Ng, L.C., Naylor, C.E., DeCaen, P.G., Wallace, B.A.(2017) Nat Commun 8: 14205-14205
- PubMed: 28205548
- DOI: https://doi.org/10.1038/ncomms14205
- Primary Citation of Related Structures:
5HVD, 5HVX - PubMed Abstract:
Voltage-gated sodium channels (Navs) play essential roles in excitable tissues, with their activation and opening resulting in the initial phase of the action potential. The cycling of Navs through open, closed and inactivated states, and their closely choreographed relationships with the activities of other ion channels lead to exquisite control of intracellular ion concentrations in both prokaryotes and eukaryotes. Here we present the 2.45 Å resolution crystal structure of the complete NavMs prokaryotic sodium channel in a fully open conformation. A canonical activated conformation of the voltage sensor S4 helix, an open selectivity filter leading to an open activation gate at the intracellular membrane surface and the intracellular C-terminal domain are visible in the structure. It includes a heretofore unseen interaction motif between W77 of S3, the S4-S5 interdomain linker, and the C-terminus, which is associated with regulation of opening and closing of the intracellular gate.
- Institute of Structural and Molecular Biology, Birkbeck College, University of London, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: