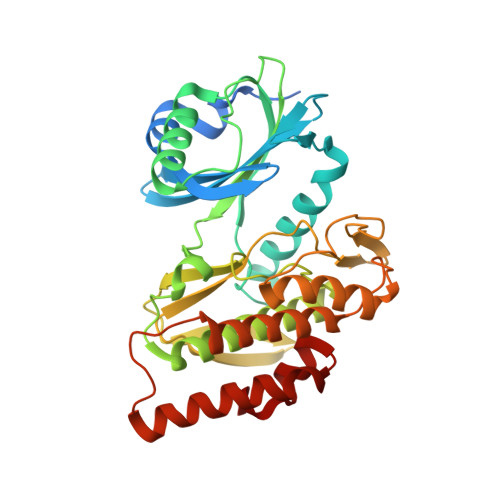
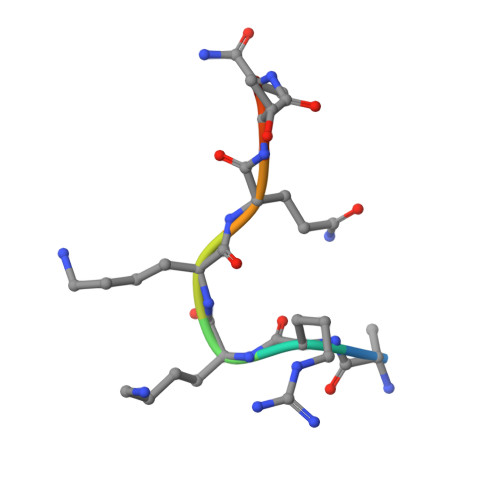
Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Lavogina, D., Kestav, K., Chaikuad, A., Heroven, C., Knapp, S., Uri, A.(2016) Acta Crystallogr F Struct Biol Commun 72: 339-345
- PubMed: 27139824
- DOI: https://doi.org/10.1107/S2053230X16004611
- Primary Citation of Related Structures:
5HTB, 5HTC - PubMed Abstract:
Haspin is a mitotic protein kinase that is responsible for the phosphorylation of Thr3 of histone H3, thereby creating a recognition motif for docking of the chromosomal passenger complex that is crucial for the progression of cell division. Here, two high-resolution models of haspin with previously reported inhibitors consisting of an ATP analogue and a histone H3(1-7) peptide analogue are presented. The structures of the complexes confirm the bisubstrate character of the inhibitors by revealing the signature binding modes of the moieties targeting the ATP-binding site and the protein substrate-binding site of the kinase. This is the first structural model of a bisubstrate inhibitor targeting haspin. The presented structural data represent a model for the future development of more specific haspin inhibitors.
- Institute of Chemistry, University of Tartu, Ravila 14A, 50411 Tartu, Estonia.
Organizational Affiliation: