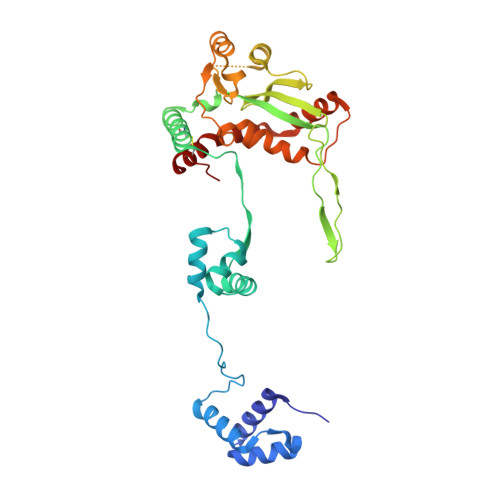
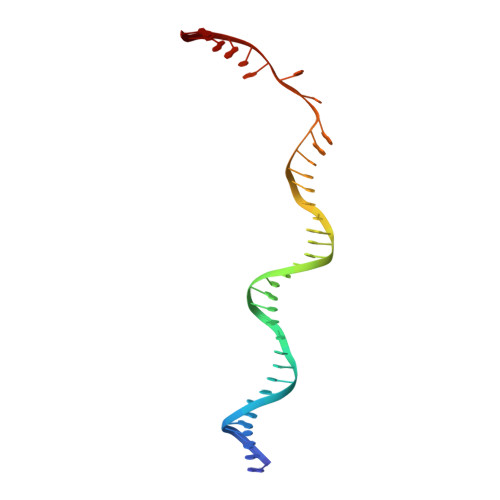
A bend, flip and trap mechanism for transposon integration.
Morris, E.R., Grey, H., McKenzie, G., Jones, A.C., Richardson, J.M.(2016) Elife 5
- PubMed: 27223327
- DOI: https://doi.org/10.7554/eLife.15537
- Primary Citation of Related Structures:
5HOO - PubMed Abstract:
Cut-and-paste DNA transposons of the mariner/Tc1 family are useful tools for genome engineering and are inserted specifically at TA target sites. A crystal structure of the mariner transposase Mos1 (derived from Drosophila mauritiana), in complex with transposon ends covalently joined to target DNA, portrays the transposition machinery after DNA integration. It reveals severe distortion of target DNA and flipping of the target adenines into extra-helical positions. Fluorescence experiments confirm dynamic base flipping in solution. Transposase residues W159, R186, F187 and K190 stabilise the target DNA distortions and are required for efficient transposon integration and transposition in vitro. Transposase recognises the flipped target adenines via base-specific interactions with backbone atoms, offering a molecular basis for TA target sequence selection. Our results will provide a template for re-designing mariner/Tc1 transposases with modified target specificities.
- Institute of Quantitative Biology, Biochemistry and Biotechnology, University of Edinburgh, Edinburgh, United Kingdom.
Organizational Affiliation: