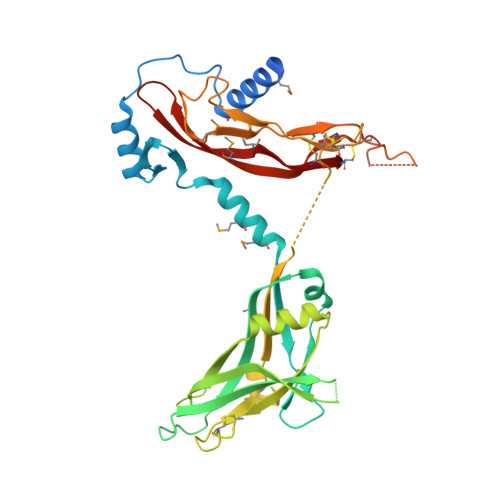
Structure and activation of pro-activin A.
Wang, X., Fischer, G., Hyvonen, M.(2016) Nat Commun 7: 12052-12052
- PubMed: 27373274
- DOI: https://doi.org/10.1038/ncomms12052
- Primary Citation of Related Structures:
5HLY, 5HLZ - PubMed Abstract:
Activins are growth factors with multiple roles in the development and homeostasis. Like all TGF-β family of growth factors, activins are synthesized as large precursors from which mature dimeric growth factors are released proteolytically. Here we have studied the activation of activin A and determined crystal structures of the unprocessed precursor and of the cleaved pro-mature complex. Replacing the natural furin cleavage site with a HRV 3C protease site, we show how the protein gains its bioactivity after proteolysis and is as active as the isolated mature domain. The complex remains associated in conditions used for biochemical analysis with a dissociation constant of 5 nM, but the pro-domain can be actively displaced from the complex by follistatin. Our high-resolution structures of pro-activin A share features seen in the pro-TGF-β1 and pro-BMP-9 structures, but reveal a new oligomeric arrangement, with a domain-swapped, cross-armed conformation for the protomers in the dimeric protein.
- Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK.
Organizational Affiliation: