Exploring the role of the alpha-carboxyphosphonate moiety in the HIV-RT activity of alpha-carboxy nucleoside phosphonates.
Mullins, N.D., Maguire, N.M., Ford, A., Das, K., Arnold, E., Balzarini, J., Maguire, A.R.(2016) Org Biomol Chem 14: 2454-2465
- PubMed: 26813581
- DOI: https://doi.org/10.1039/c5ob02507a
- Primary Citation of Related Structures:
5HLF - PubMed Abstract:
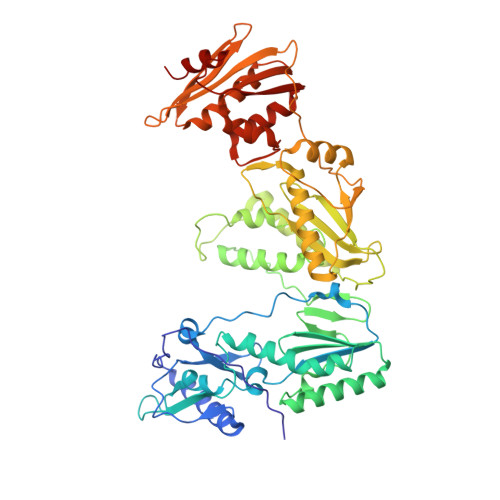
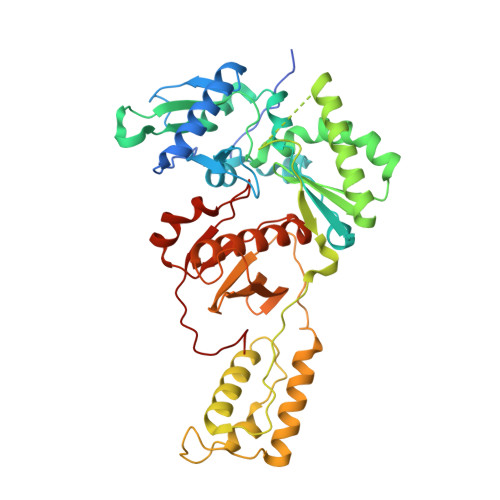
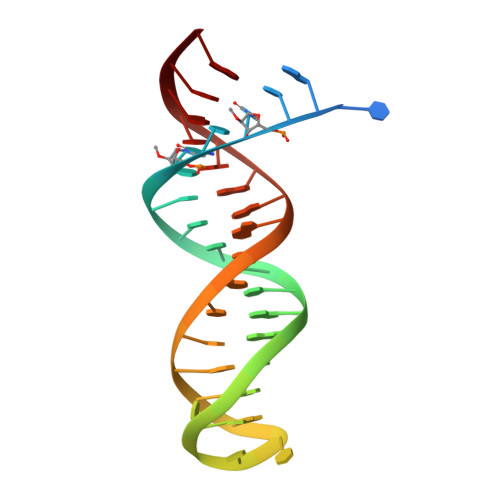
As α-carboxy nucleoside phosphonates (α-CNPs) have demonstrated a novel mode of action of HIV-1 reverse transcriptase inhibition, structurally related derivatives were synthesized, namely the malonate 2, the unsaturated and saturated bisphosphonates 3 and 4, respectively and the amide 5. These compounds were evaluated for inhibition of HIV-1 reverse transcriptase in cell-free assays. The importance of the α-carboxy phosphonoacetic acid moiety for achieving reverse transcriptase inhibition, without the need for prior phosphorylation, was confirmed. The malonate derivative 2 was less active by two orders of magnitude than the original α-CNPs, while displaying the same pattern of kinetic behavior; interestingly the activity resides in the “L”-enantiomer of 2, as seen with the earlier series of α-CNPs. A crystal structure with an RT/DNA complex at 2.95 Å resolution revealed the binding of the “L”-enantiomer of 2, at the polymerase active site with a weaker metal ion chelation environment compared to 1a (T-α-CNP) which may explain the lower inhibitory activity of 2.
- Department of Chemistry, Analytical and Biological Chemistry Research Facility, Synthesis and Solid State Pharmaceutical Centre, University College, Cork, Ireland. a.maguire@ucc.ie.
Organizational Affiliation: