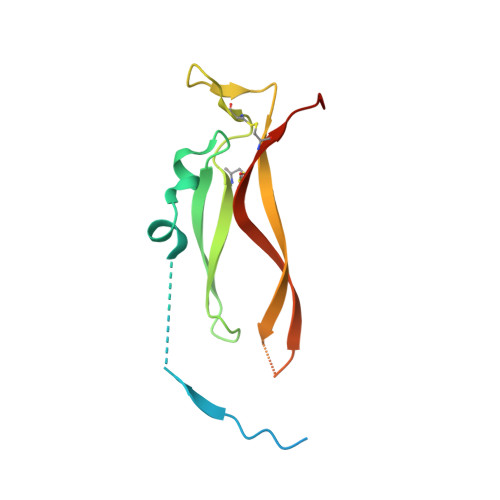
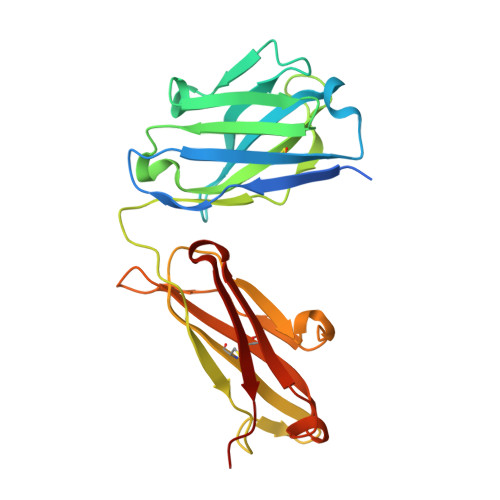
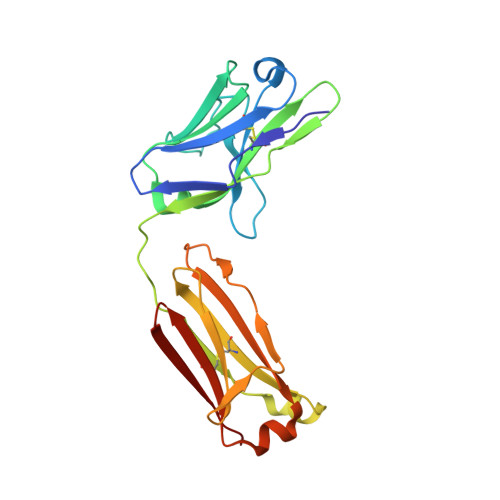
Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Liu, S., Dakin, L.A., Xing, L., Withka, J.M., Sahasrabudhe, P.V., Li, W., Banker, M.E., Balbo, P., Shanker, S., Chrunyk, B.A., Guo, Z., Chen, J.M., Young, J.A., Bai, G., Starr, J.T., Wright, S.W., Bussenius, J., Tan, S., Gopalsamy, A., Lefker, B.A., Vincent, F., Jones, L.H., Xu, H., Hoth, L.R., Geoghegan, K.F., Qiu, X., Bunnage, M.E., Thorarensen, A.(2016) Sci Rep 6: 30859-30859
- PubMed: 27527709
- DOI: https://doi.org/10.1038/srep30859
- Primary Citation of Related Structures:
5HI3, 5HI4, 5HI5 - PubMed Abstract:
Interleukin-17A (IL-17A) is a principal driver of multiple inflammatory and immune disorders. Antibodies that neutralize IL-17A or its receptor (IL-17RA) deliver efficacy in autoimmune diseases, but no small-molecule IL-17A antagonists have yet progressed into clinical trials. Investigation of a series of linear peptide ligands to IL-17A and characterization of their binding site has enabled the design of novel macrocyclic ligands that are themselves potent IL-17A antagonists.
- Worldwide Medicinal Chemistry, Pfizer Worldwide R&D, Eastern Point Road, Groton, CT 06340, USA.
Organizational Affiliation: