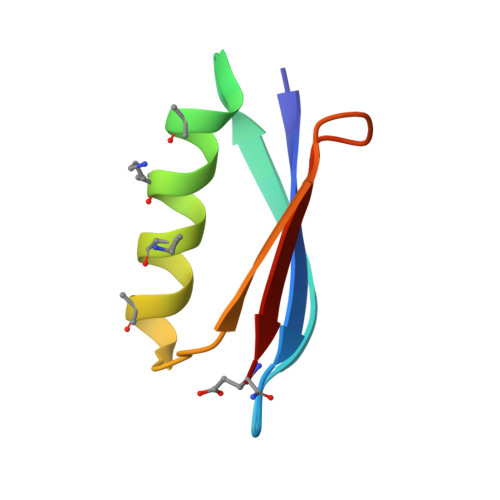
Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold.
Tavenor, N.A., Reinert, Z.E., Lengyel, G.A., Griffith, B.D., Horne, W.S.(2016) Chem Commun (Camb) 52: 3789-3792
- PubMed: 26853882
- DOI: https://doi.org/10.1039/c6cc00273k
- Primary Citation of Related Structures:
5HFY, 5HG2, 5HI1 - PubMed Abstract:
We report here the comparison of five classes of unnatural amino acid building blocks for their ability to be accommodated into an α-helix in a protein tertiary fold context. High-resolution structural characterization and analysis of folding thermodynamics yield new insights into the relationship between backbone composition and folding energetics in α-helix mimetics and suggest refined design rules for engineering the backbones of natural sequences.
- Department of Chemistry, University of Pittsburgh, Pittsburgh, PA 15260, USA. horne@pitt.edu.
Organizational Affiliation: