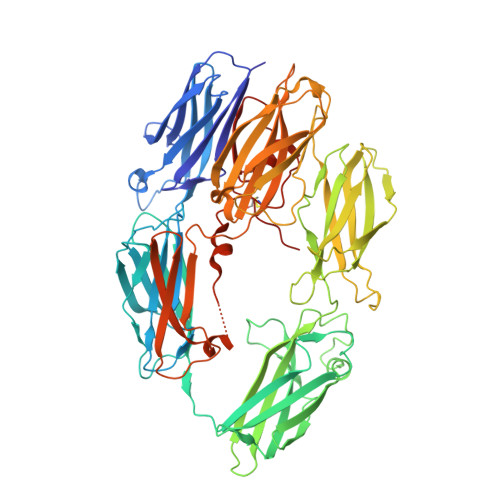
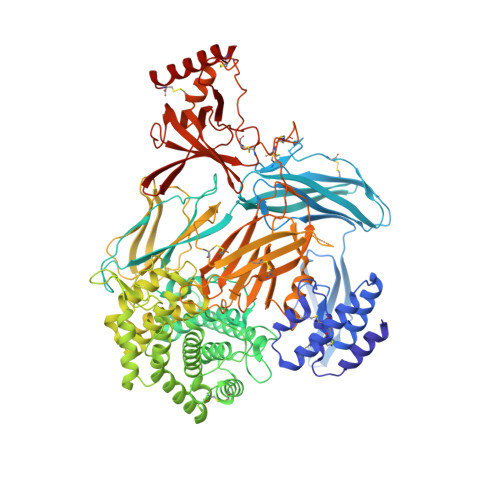
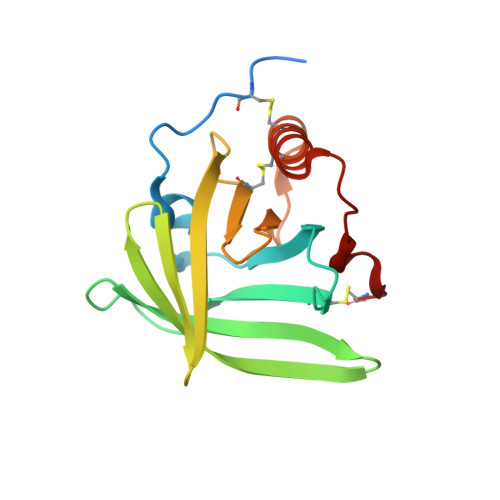
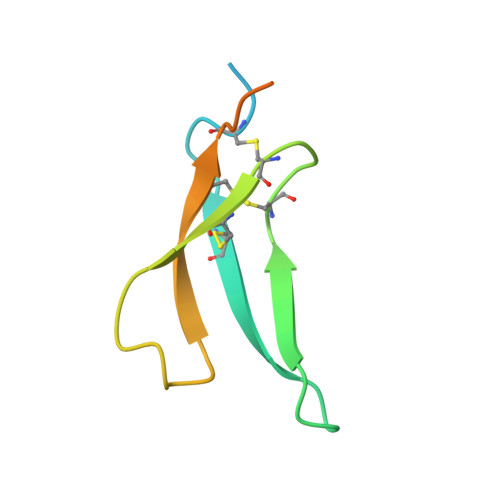
Structural basis for therapeutic inhibition of complement C5.
Jore, M.M., Johnson, S., Sheppard, D., Barber, N.M., Li, Y.I., Nunn, M.A., Elmlund, H., Lea, S.M.(2016) Nat Struct Mol Biol 23: 378-386
- PubMed: 27018802
- DOI: https://doi.org/10.1038/nsmb.3196
- Primary Citation of Related Structures:
5HCC, 5HCD, 5HCE, 5IEC - PubMed Abstract:
Activation of complement C5 generates the potent anaphylatoxin C5a and leads to pathogen lysis, inflammation and cell damage. The therapeutic potential of C5 inhibition has been demonstrated by eculizumab, one of the world's most expensive drugs. However, the mechanism of C5 activation by C5 convertases remains elusive, thus limiting development of therapeutics. Here we identify and characterize a new protein family of tick-derived C5 inhibitors. Structures of C5 in complex with the new inhibitors, the phase I and phase II inhibitor OmCI, or an eculizumab Fab reveal three distinct binding sites on C5 that all prevent activation of C5. The positions of the inhibitor-binding sites and the ability of all three C5-inhibitor complexes to competitively inhibit the C5 convertase conflict with earlier steric-inhibition models, thus suggesting that a priming event is needed for activation.
- Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.
Organizational Affiliation: