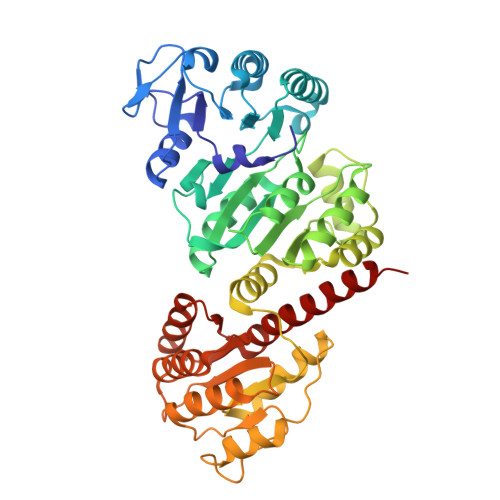
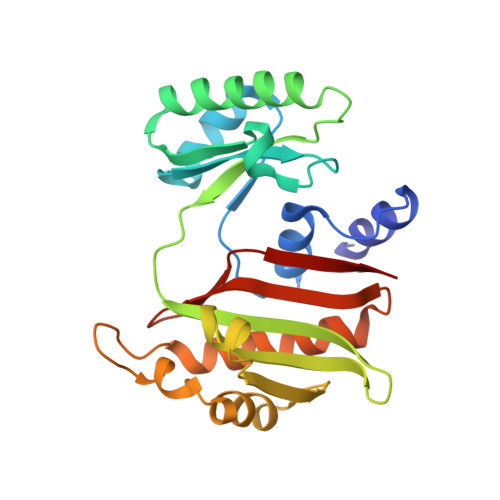
Structure of NDP-forming Acetyl-CoA synthetase ACD1 reveals a large rearrangement for phosphoryl transfer.
Weie, R.H., Faust, A., Schmidt, M., Schonheit, P., Scheidig, A.J.(2016) Proc Natl Acad Sci U S A 113: E519-E528
- PubMed: 26787904
- DOI: https://doi.org/10.1073/pnas.1518614113
- Primary Citation of Related Structures:
4XYL, 4XYM, 4XZ3, 4Y8V, 4YAJ, 4YAK, 4YB8, 4YBZ, 5HBR - PubMed Abstract:
The NDP-forming acyl-CoA synthetases (ACDs) catalyze the conversion of various CoA thioesters to the corresponding acids, conserving their chemical energy in form of ATP. The ACDs are the major energy-conserving enzymes in sugar and peptide fermentation of hyperthermophilic archaea. They are considered to be primordial enzymes of ATP synthesis in the early evolution of life. We present the first crystal structures, to our knowledge, of an ACD from the hyperthermophilic archaeon Candidatus Korachaeum cryptofilum. These structures reveal a unique arrangement of the ACD subunits alpha and beta within an α2β2-heterotetrameric complex. This arrangement significantly differs from other members of the superfamily. To transmit an activated phosphoryl moiety from the Ac-CoA binding site (within the alpha subunit) to the NDP-binding site (within the beta subunit), a distance of 51 Å has to be bridged. This transmission requires a larger rearrangement within the protein complex involving a 21-aa-long phosphohistidine-containing segment of the alpha subunit. Spatial restraints of the interaction of this segment with the beta subunit explain the necessity for a second highly conserved His residue within the beta subunit. The data support the proposed four-step reaction mechanism of ACDs, coupling acyl-CoA thioesters with ATP synthesis. Furthermore, the determined crystal structure of the complex with bound Ac-CoA allows first insight, to our knowledge, into the determinants for acyl-CoA substrate specificity. The composition and size of loops protruding into the binding pocket of acyl-CoA are determined by the individual arrangement of the characteristic subdomains.
- Zoological Institute-Structural Biology, Kiel University, 24118 Kiel, Germany; Centre for Biotechnology and Biomedicine, Structure Analysis of Biopolymers, Leipzig University, 04103 Leipzig, Germany;
Organizational Affiliation: