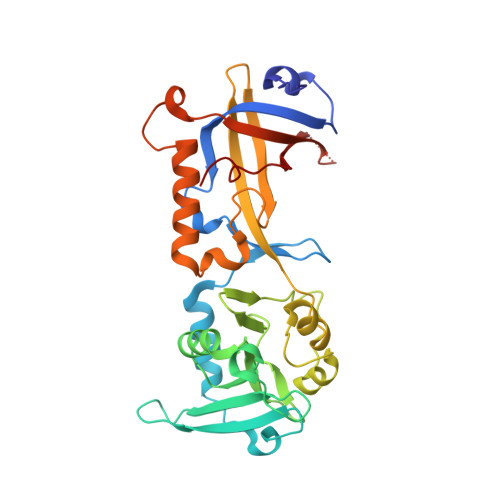
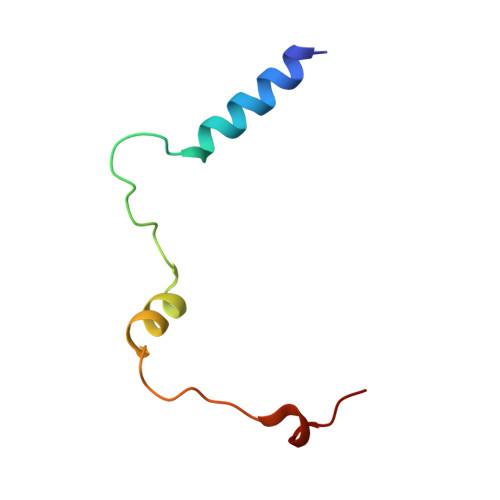
Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Chen, C., Gu, P., Wu, J., Chen, X., Niu, S., Sun, H., Wu, L., Li, N., Peng, J., Shi, S., Fan, C., Huang, M., Wong, C.C., Gong, Q., Kumar-Sinha, C., Zhang, R., Pusztai, L., Rai, R., Chang, S., Lei, M.(2017) Nat Commun 8: 14929-14929
- PubMed: 28393832
- DOI: https://doi.org/10.1038/ncomms14929
- Primary Citation of Related Structures:
5H65 - PubMed Abstract:
Mammalian shelterin proteins POT1 and TPP1 form a stable heterodimer that protects chromosome ends and regulates telomerase-mediated telomere extension. However, how POT1 interacts with TPP1 remains unknown. Here we present the crystal structure of the C-terminal portion of human POT1 (POT1C) complexed with the POT1-binding motif of TPP1. The structure shows that POT1C contains two domains, a third OB fold and a Holliday junction resolvase-like domain. Both domains are essential for binding to TPP1. Notably, unlike the heart-shaped structure of ciliated protozoan Oxytricha nova TEBPα-β complex, POT1-TPP1 adopts an elongated V-shaped conformation. In addition, we identify several missense mutations in human cancers that disrupt the POT1C-TPP1 interaction, resulting in POT1 instability. POT1C mutants that bind TPP1 localize to telomeres but fail to repress a DNA damage response and inappropriate repair by A-NHEJ. Our results reveal that POT1 C terminus is essential to prevent initiation of genome instability permissive for tumorigenesis.
- National Center for Protein Science Shanghai, State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, 333 Haike Road, Shanghai 201210, China.
Organizational Affiliation: