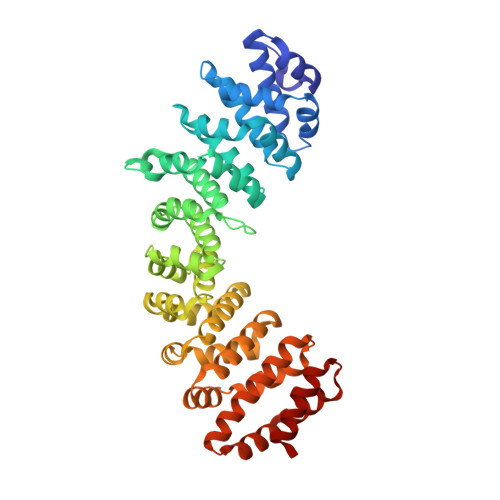
Structural insights into the nuclear import of the histone acetyltransferase males-absent-on-the-first by importin alpha 1
Zheng, W., Wang, R., Liu, X., Tian, S., Yao, B., Chen, A., Jin, S., Li, Y.(2018) Traffic 19: 19-28
- PubMed: 28991411
- DOI: https://doi.org/10.1111/tra.12534
- Primary Citation of Related Structures:
5H43 - PubMed Abstract:
The histone acetyltransferase males-absent-on-the-first (MOF) acetylates the histone H4, a modification important for many biological processes, including chromatin organization, transcriptional regulation, DNA replication, recombination and repair, as well as autophagy. Depletion of MOF induces serious consequences because of the reduction of histone acetylation, such as nuclear morphological defects and cancer. Despite the critical roles of MOF in the nucleus, the structural or functional mechanisms of the nucleocytoplasmic transport of MOF remain elusive. Here, we identified novel importin α1-specific nuclear localization signals (NLSs) in the N-terminal of human MOF. The crystal structure of MOF NLSs in complex with importin α1 further revealed a unique binding mode of MOF, with two independent NLSs binding to importin α1 major and minor sites, respectively. The second NLS of MOF displays an unexpected α-helical conformation in the C-terminus, with more extensive contacts with importin α1 not limited in the minor site. Mutations of the key residues on MOF and importin α1 lead to the reduction of their interaction as well as the nuclear import of MOF, revealing an essential role of NLS2 of MOF in interacting with importin α1 minor site. Taken together, we provide structural mechanisms underlying the nucleocytoplasmic transport of MOF, which will be of great importance in understanding the functional regulation of MOF in various biological processes.
- State Key Laboratory of Cellular Stress Biology, Innovation Center for Cell Signaling Network, School of Life Sciences, Xiamen University, Fujian, China.
Organizational Affiliation: