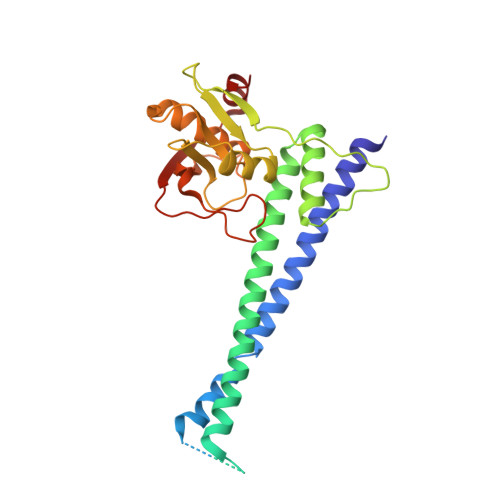
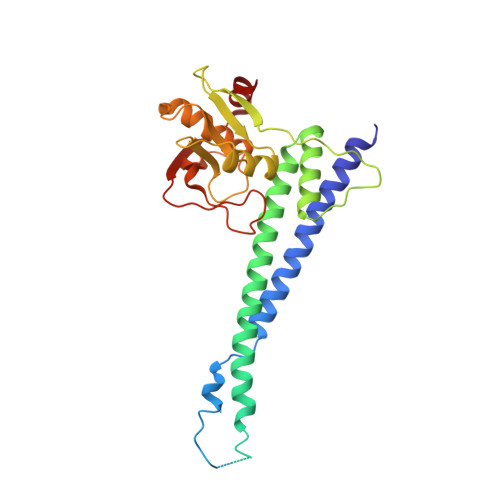
Structural and functional analysis of an anchorless fibronectin-binding protein FBPS from Gram-positive bacterium Streptococcus suis
Musyoki, A.M., Shi, Z., Xuan, C., Lu, G., Qi, J., Gao, F., Zheng, B., Zhang, Q., Li, Y., Haywood, J., Liu, C., Yan, J., Shi, Y., Gao, G.F.(2016) Proc Natl Acad Sci U S A 113: 13869-13874
- PubMed: 27834729
- DOI: https://doi.org/10.1073/pnas.1608406113
- Primary Citation of Related Structures:
5H3W, 5H3X - PubMed Abstract:
The anchorless fibronectin-binding proteins (FnBPs) are a group of important virulence factors for which the structures are not available and the functions are not well defined. In this study we performed comprehensive studies on a prototypic member of this group: the fibronectin-/fibrinogen-binding protein from Streptococcus suis (FBPS). The structures of the N- and C-terminal halves (FBPS-N and FBPS-C), which together cover the full-length protein in sequence, were solved at a resolution of 2.1 and 2.6 Å, respectively, and each was found to be composed of two domains with unique folds. Furthermore, we have elucidated the organization of these domains by small-angle X-ray scattering. We further showed that the fibronectin-binding site is located in FBPS-C and that FBPS promotes the adherence of S suis to host cells by attaching the bacteria via FBPS-N. Finally, we demonstrated that FBPS functions both as an adhesin, promoting S suis attachment to host cells, and as a bacterial factor, activating signaling pathways via β1 integrin receptors to induce chemokine production.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: