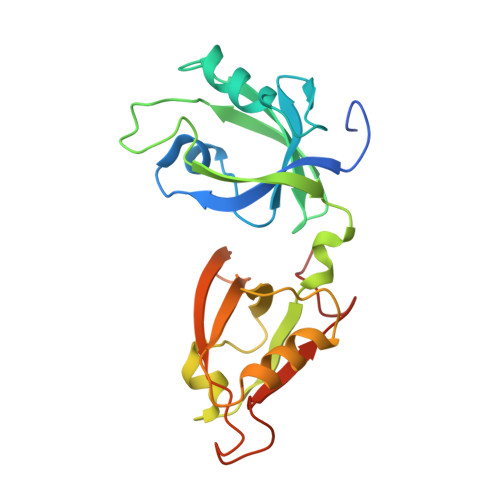
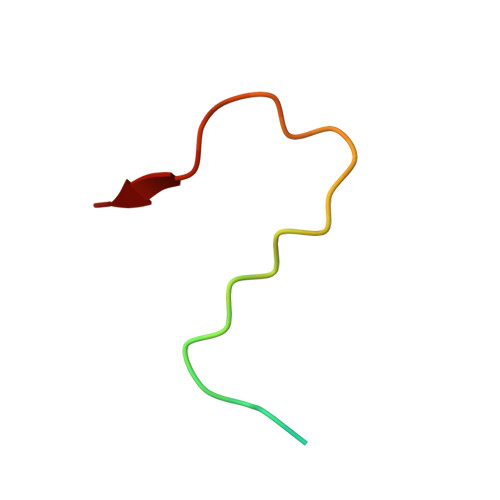
Structural Basis for the Interaction between Golgi Reassembly-stacking Protein GRASP55 and Golgin45
Zhao, J., Li, B., Huang, X., Morelli, X., Shi, N.(2017) J Biological Chem 292: 2956-2965
- PubMed: 28049725
- DOI: https://doi.org/10.1074/jbc.M116.765990
- Primary Citation of Related Structures:
5H3J - PubMed Abstract:
Golgin45 is required for normal Golgi structure and the transportation of protein from the ER. It forms a specific complex with GRASP55 in vivo Little is known regarding the molecular details of this interaction and its structural role in stacking of the Golgi complex. Here, we present the crystal structure of the GRASP domains of GRASP55 in complex with the Golgin45 C-terminal peptide, determined at 1.33 Å resolution. Similar to the structure of GRASP65 bound to GM130 reported recently, this structure reveals more than one interacting site and involves both PDZ1 and PDZ2 domains of the GRASP simultaneously. The C-terminal peptides of Golgin45 and GM130 present a conserved PDZ domain binding motif sequence and recognize the canonical PDZ-peptide binding groove of the PDZ1 domains of GRASP55 and GRASP65. A main difference in this recognition process resides in a structural rearrangement of GRASP65-GM130 that does not occur for the GRASP55-Golgin45 complex. The binding site at the cleft between the PDZ1 and PDZ2 domains of GRASP65 is dominated by hydrophobic interactions with GM130 that are not observed in the GRASP55-Golgin45 complex. In addition, a unique zinc finger structure is revealed in the GRASP55-Golgin45 complex crystal structure. Mutagenesis experiments support these structural observations and demonstrate that two of these sites are required to form a stable complex. Finally, a novel Golgi stacking model is proposed according to these structural findings.
- From the State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fuzhou 350002, China and.
Organizational Affiliation: