Crystal structure of FM329, a recombinant Fab adopted from cetuximab
Sim, D.W., Kim, J.H., Kim, Y.P., Won, H.S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FM329 light chain | 215 | Homo sapiens/Mus musculus xenograft | Mutation(s): 0 | 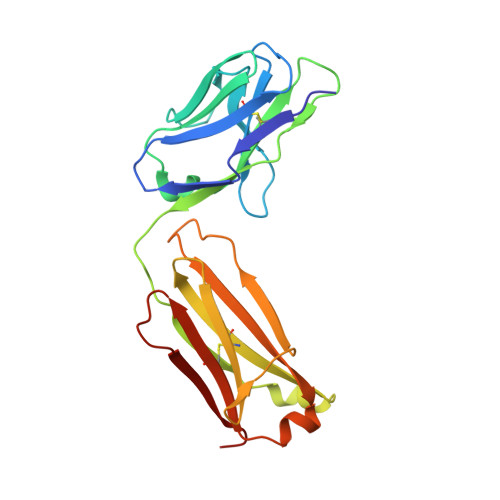 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FM329 heavy chain | 224 | Homo sapiens/Mus musculus xenograft | Mutation(s): 0 | 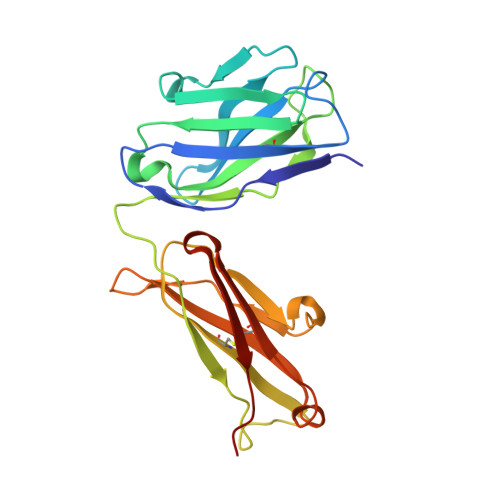 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.521 | α = 90 |
| b = 68.031 | β = 101.84 |
| c = 97.78 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data processing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Korea Small and Medium Business Administration | Korea, Republic Of | C0333276 |