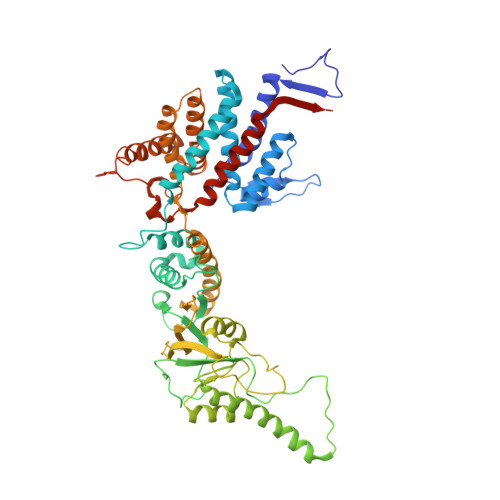
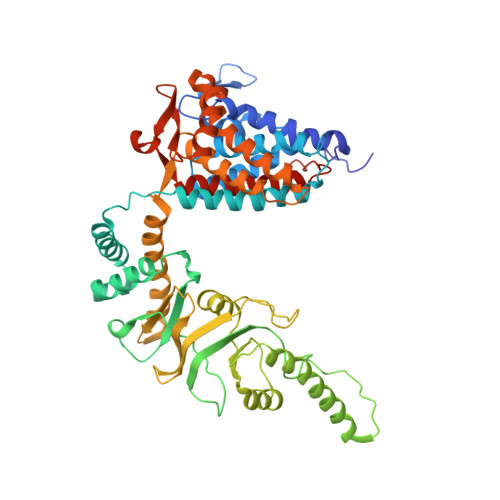
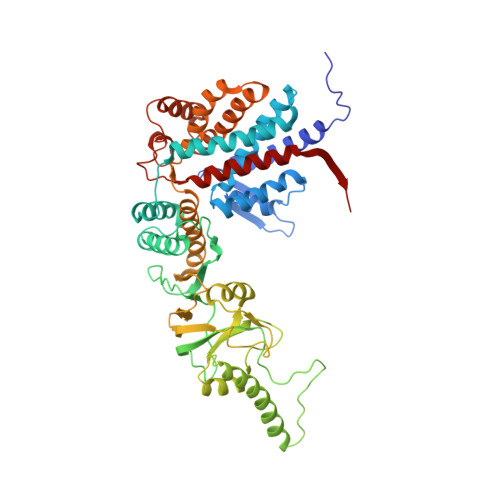
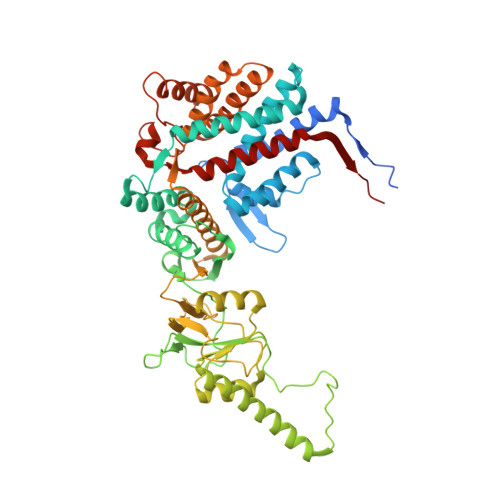
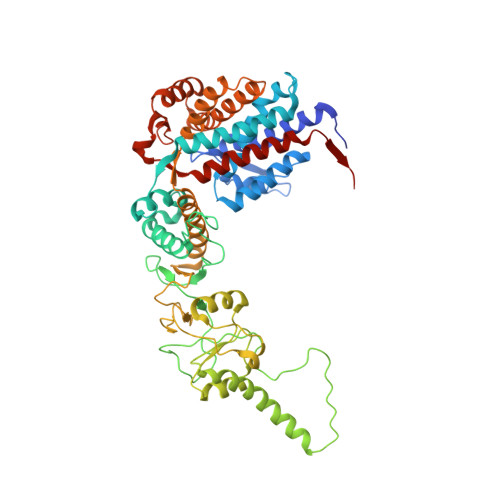
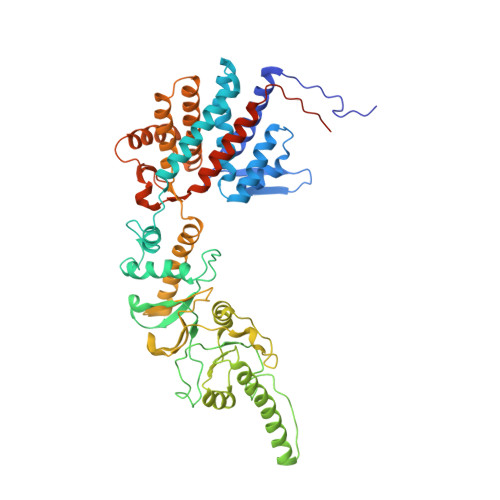
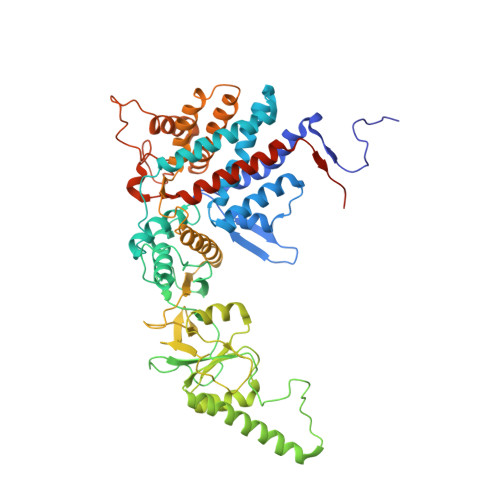
Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Zang, Y., Jin, M., Wang, H., Cui, Z., Kong, L., Liu, C., Cong, Y.(2016) Nat Struct Mol Biol 23: 1083-1091
- PubMed: 27775711
- DOI: https://doi.org/10.1038/nsmb.3309
- Primary Citation of Related Structures:
5GW4, 5GW5 - PubMed Abstract:
The eukaryotic chaperonin TRiC (or CCT) assists in the folding of 10% of cytosolic proteins. Here we present two cryo-EM structures of Saccharomyces cerevisiae TRiC in a newly identified nucleotide partially preloaded (NPP) state and in the ATP-bound state, at 4.7-Å and 4.6-Å resolution, respectively. Through inner-subunit eGFP tagging, we identified the subunit locations in open-state TRiC and found that the CCT2 subunit pair forms an unexpected Z shape. ATP binding induces a dramatic conformational change on the CCT2 side, thereby suggesting that CCT2 plays an essential role in TRiC allosteric cooperativity. Our structural and biochemical data reveal a staggered ATP binding mechanism of TRiC with preloaded nucleotide on the CCT6 side of NPP-TRiC and demonstrate that TRiC has evolved into a complex that is structurally divided into two sides. This work offers insight into how the TRiC nucleotide cycle coordinates with its mechanical cycle in preparing folding intermediates for further productive folding.
- National Center for Protein Science Shanghai, State Key Laboratory of Molecular Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: