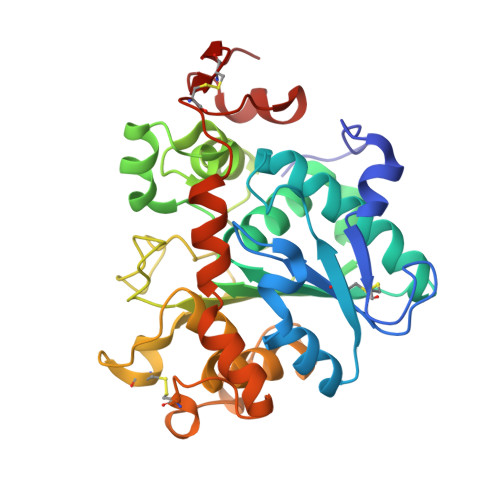
Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Park, K., Kim, S., Park, J., Joe, S., Min, B., Oh, J., Song, J., Park, S.Y., Park, S., Lee, H.(2016) ACS Catal 6: 7458-7465