Potent Antitumor Activities and Structure Basis of the Chiral beta-Lactam Bridged Analogue of Combretastatin A-4 Binding to Tubulin.
Zhou, P., Liu, Y., Zhou, L., Zhu, K., Feng, K., Zhang, H., Liang, Y., Jiang, H., Luo, C., Liu, M., Wang, Y.(2016) J Med Chem 59: 10329-10334
- PubMed: 27805821
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01268
- Primary Citation of Related Structures:
5GON - PubMed Abstract:
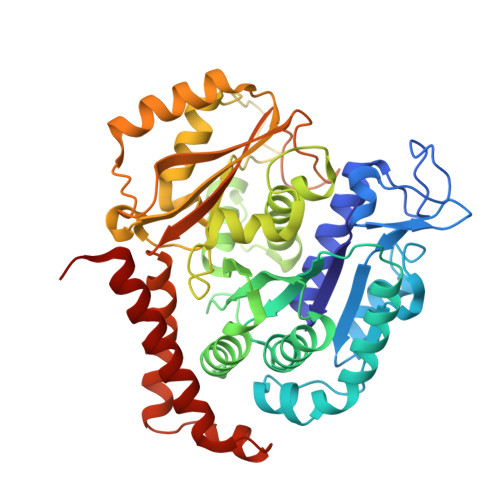
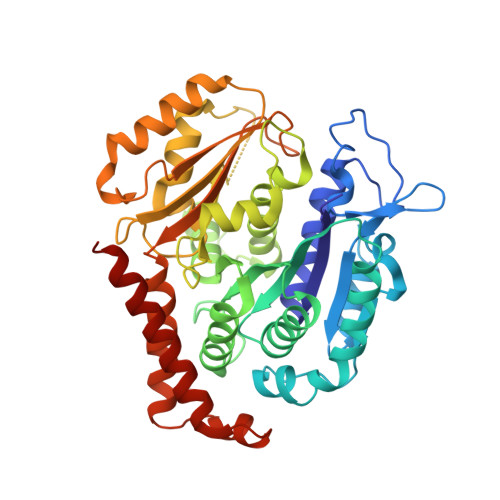
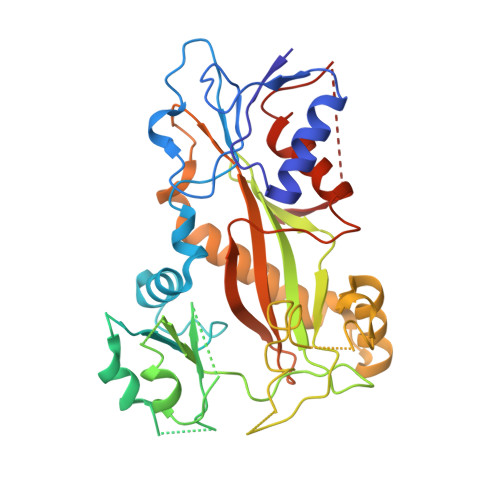
A series of chiral β-lactam bridged analogues (3-substituted 1,4-diaryl-2-azetidinones) of combretastatin A-4 (CA-4) were synthesized asymmetrically, and their antitumor activities were evaluated in vitro and in vivo. The cocrystal structure of tubulin in complex with compound 9 was determined by X-ray crystallography, which showed that 9 binds to the same site as colchicine with similar binding mode, and the absolute configuration of its C-4 was first identified and demonstrated to be critically important for their antiproliferative activities.
- School of Pharmacy, Fudan University , Shanghai 201203, China.
Organizational Affiliation: