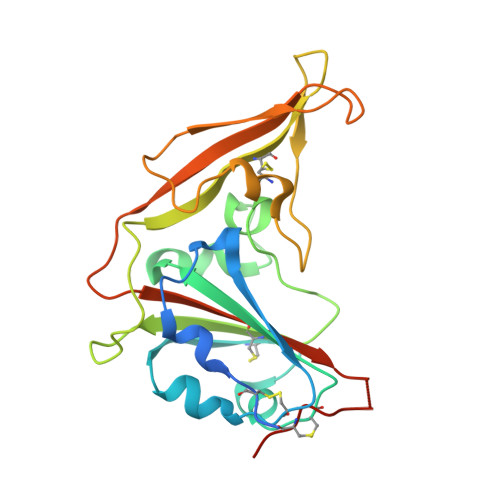
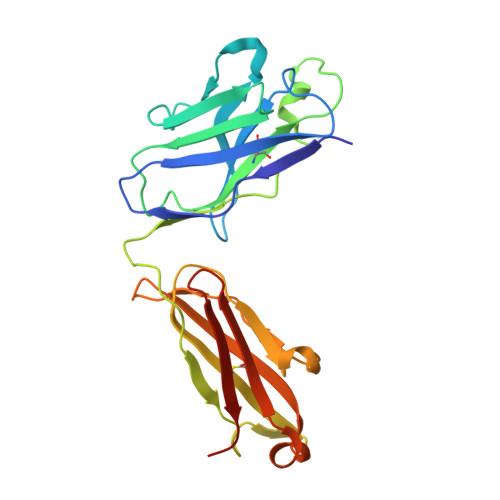
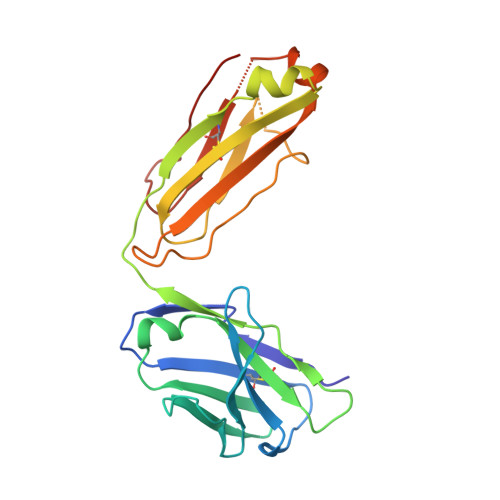
Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
Chen, Z., Bao, L., Chen, C., Zou, T., Xue, Y., Li, F., Lv, Q., Gu, S., Gao, X., Cui, S., Wang, J., Qin, C., Jin, Q.(2017) J Infect Dis 215: 1807-1815
- PubMed: 28472421
- DOI: https://doi.org/10.1093/infdis/jix209
- Primary Citation of Related Structures:
5GMQ - PubMed Abstract:
Middle East respiratory syndrome coronavirus (MERS-CoV) infection in humans is highly lethal, with a fatality rate of 35%. New prophylactic and therapeutic strategies to combat human infections are urgently needed. We isolated a fully human neutralizing antibody, MCA1, from a human survivor. The antibody recognizes the receptor-binding domain of MERS-CoV S glycoprotein and interferes with the interaction between viral S and the human cellular receptor human dipeptidyl peptidase 4 (DPP4). To our knowledge, this study is the first to report a human neutralizing monoclonal antibody that completely inhibits MERS-CoV replication in common marmosets. Monotherapy with MCA1 represents a potential alternative treatment for human infections with MERS-CoV worthy of evaluation in clinical settings.
- MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College.
Organizational Affiliation: