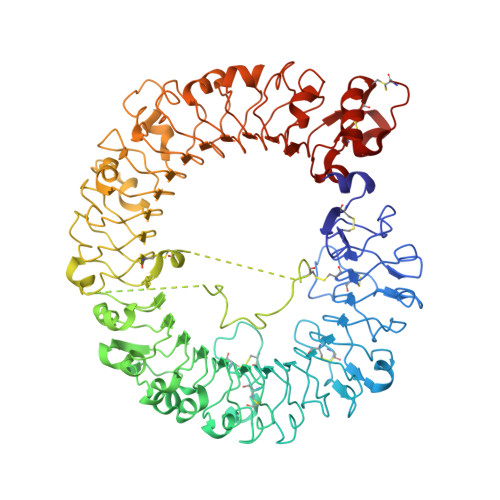
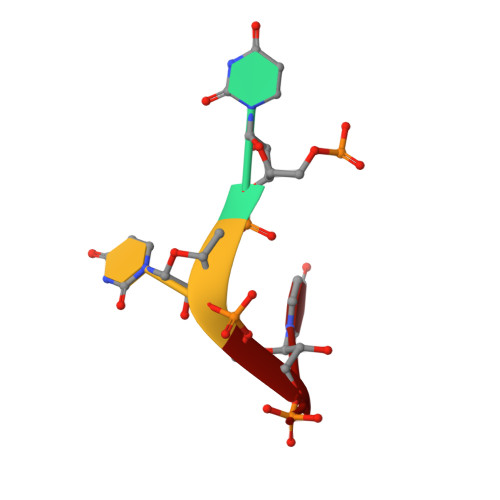
Structural Analysis Reveals that Toll-like Receptor 7 Is a Dual Receptor for Guanosine and Single-Stranded RNA
Zhang, Z., Ohto, U., Shibata, T., Krayukhina, E., Taoka, M., Yamauchi, Y., Tanji, H., Isobe, T., Uchiyama, S., Miyake, K., Shimizu, T.(2016) Immunity 45: 737-748
- PubMed: 27742543
- DOI: https://doi.org/10.1016/j.immuni.2016.09.011
- Primary Citation of Related Structures:
5GMF, 5GMG, 5GMH - PubMed Abstract:
Toll-like receptor 7 (TLR7) is a single-stranded RNA (ssRNA) sensor in innate immunity and also responds to guanosine and chemical ligands, such as imidazoquinoline compounds. However, TLR7 activation mechanism by these ligands remain largely unknown. Here, we generated crystal structures of three TLR7 complexes, and found that all formed an activated m-shaped dimer with two ligand-binding sites. The first site conserved in TLR7 and TLR8 was used for small ligand-binding essential for its activation. The second site spatially distinct from that of TLR8 was used for a ssRNA-binding that enhanced the affinity of the first-site ligands. The first site preferentially recognized guanosine and the second site specifically bound to uridine moieties in ssRNA. Our structural, biochemical, and mutagenesis studies indicated that TLR7 is a dual receptor for guanosine and uridine-containing ssRNA. Our findings have important implications for understanding of TLR7 function, as well as for therapeutic manipulation of TLR7 activation.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: