Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
Ryu, B.H., Ngo, T.D., Kim, B.Y., Yoo, W.K., Lee, E.J., Lee, S.J., Kim, K.K., Kim, T.D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Esterase A | 386 | Caulobacter vibrioides CB15 | Mutation(s): 0 Gene Names: CC_0255 | 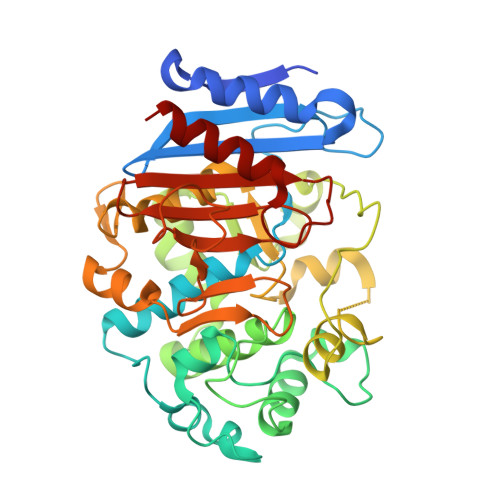 | |
UniProt | |||||
Find proteins for Q9ABH2 (Caulobacter vibrioides (strain ATCC 19089 / CIP 103742 / CB 15)) Explore Q9ABH2 Go to UniProtKB: Q9ABH2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9ABH2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.277 | α = 90 |
| b = 67.308 | β = 90 |
| c = 92.093 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| HKL-2000 | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation of Korea | Korea, Republic Of | NRF-2013K2A1A2053659 |