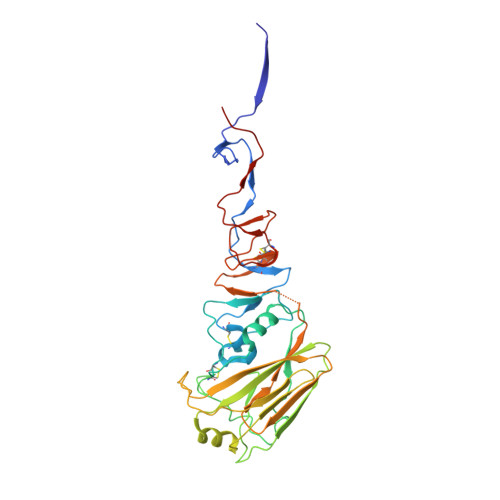
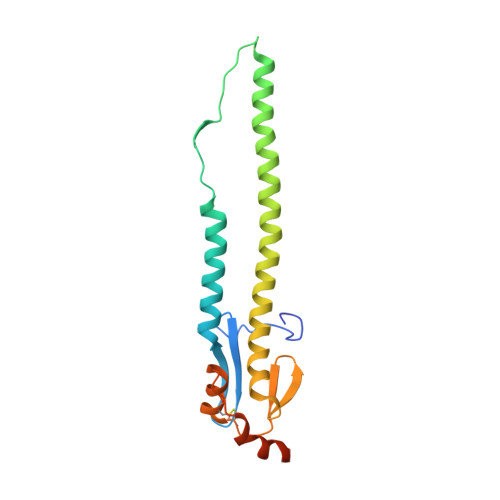
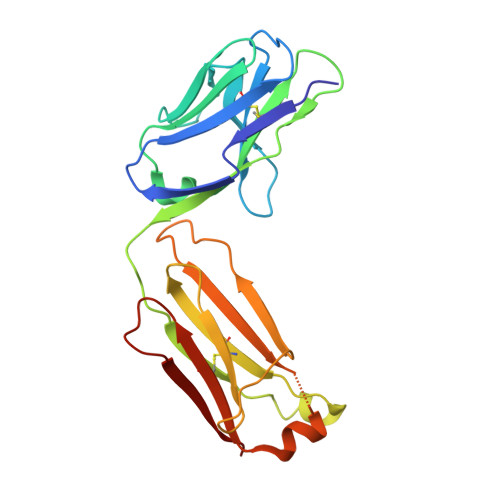
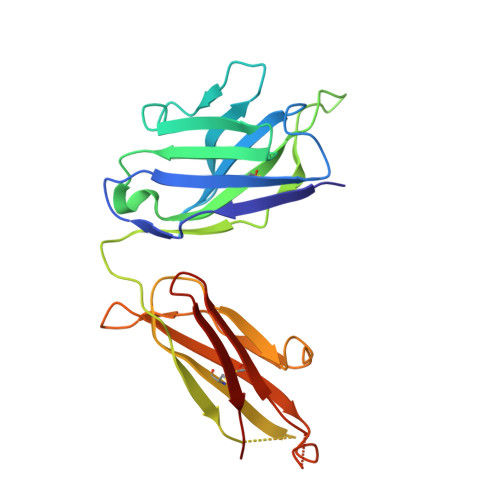
Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza A viruses
Wang, W., Sun, X., Li, Y., Su, J., Ling, Z., Zhang, T., Wang, F., Zhang, H., Chen, H., Ding, J., Sun, B.(2016) Nat Commun 7: 13577-13577
- PubMed: 27910950
- DOI: https://doi.org/10.1038/ncomms13577
- Primary Citation of Related Structures:
5GJS, 5GJT - PubMed Abstract:
As influenza A viruses remain a major threat to human health worldwide, the discovery of broadly neutralizing monoclonal antibodies that recognize conserved epitopes would facilitate the development of antibody-based therapeutic strategies. Here we report that a V H 4-4-encoded human mAb named 3E1 could neutralize H1 and H5 subtype viruses in vitro and protect mice against the H1N1 and H5N6 viruses by inhibiting the low pH-induced conformational rearrangement of haemagglutinin (HA), hence blocking membrane fusion. The crystal structures of 3E1 Fab in complex with HA of two H1N1 strains reveal that 3E1, with both heavy and light chains, binds to a conserved epitope of the HA stem region, comprising parts of the fusion peptide, the F subdomain and the outermost β-strand preceding helix A. Altogether, these data suggest the potential of 3E1 as a therapeutic drug against H1 and H5 subtype viruses.
- National Center for Protein Science Shanghai, State Key Laboratory of Molecular Biology, Center for Excellence in Molecular Cell Science, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences and University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai 200031, China.
Organizational Affiliation: